Abstract
Whether post-transcriptional regulation of gene expression controls differentiation of stem cells for tissue renewal remains unknown. Quiescent stem cells exhibit a low level of protein synthesis1, which is key to maintaining the pool of fully functional stem cells, not only in the brain but also in the bone marrow and hair follicles2,3,4,5,6. Neurons also maintain a subset of messenger RNAs in a translationally silent state, which react ‘on demand’ to intracellular and extracellular signals. This uncoupling of general availability of mRNA from translation into protein facilitates immediate responses to environmental changes and avoids excess production of proteins, which is the most energy-consuming process within the cell. However, when post-transcriptional regulation is acquired and how protein synthesis changes along the different steps of maturation are not known. Here we show that protein synthesis undergoes highly dynamic changes when stem cells differentiate to neurons in vivo. Examination of individual transcripts using RiboTag mouse models reveals that whereas stem cells translate abundant transcripts with little discrimination, translation becomes increasingly regulated with the onset of differentiation. The generation of neurogenic progeny involves translational repression of a subset of mRNAs, including mRNAs that encode the stem cell identity factors SOX2 and PAX6, and components of the translation machinery, which are enriched in a pyrimidine-rich motif. The decrease of mTORC1 activity as stem cells exit the cell cycle selectively blocks translation of these transcripts. Our results reveal a control mechanism by which the cell cycle is coupled to post-transcriptional repression of key stem cell identity factors, thereby promoting exit from stemness.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw total mRNA, ribo-IP+ and ribo-IP− sequence data have been deposited in the Gene Expression Omnibus under accession number GSE94991. Source Data for bar graphs and box plots in Figures and Extended Data Figures are provided in the online version of the paper.
References
Llorens-Bobadilla, E. et al. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340 (2015).
Fuentealba, L. C. et al. embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655 (2015).
Furutachi, S. et al. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 18, 657–665 (2015).
Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016).
Signer, R. A. J., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014).
Bonaguidi, M. A. et al. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155 (2011).
Codega, P. et al. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 82, 545–559 (2014).
Mich, J. K. et al. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife 3, 894 (2014).
Bayraktar, O. A., Fuentealba, L. C., Alvarez-Buylla, A. & Rowitch, D. H. Astrocyte development and heterogeneity. Cold Spring Harb. Perspect. Biol. 7, a020362 (2015).
Liu, J., Xu, Y., Stoleru, D. & Salic, A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl Acad. Sci. USA 109, 413–418 (2012).
Sanz, E. et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl Acad. Sci. USA 106, 13939–13944 (2009).
Sanz, E. et al. RiboTag analysis of actively translated mRNAs in Sertoli and Leydig cells in vivo. PLoS ONE 8, e66179 (2013).
Shigeoka, T. et al. Dynamic axonal translation in developing and mature visual circuits. Cell 166, 181–192 (2016).
Cabezas-Wallscheid, N. et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome. Stem Cells 4, 502–522 (2014).
Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA 112, 15672–15677 (2015).
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Paliouras, G. N. et al. Mammalian target of rapamycin signaling is a key regulator of the transit-amplifying progenitor pool in the adult and aging forebrain. J. Neurosci. 32, 15012–15026 (2012).
Demetriades, C., Doumpas, N. & Teleman, A. A. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799 (2014).
Thoreen, C. C. et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009).
Stumpf, C. R., Moreno, M. V., Olshen, A. B., Taylor, B. S. & Ruggero, D. The translational landscape of the mammalian cell cycle. Mol. Cell 52, 574–582 (2013).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Tahmasebi, S. et al. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell 14, 606–616 (2014).
Meyer, K. D. & Jaffrey, S. R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Costa, V. et al. mTORC1 inhibition corrects neurodevelopmental and synaptic alterations in a human stem cell model of tuberous sclerosis. Cell Reports 15, 86–95 (2016).
Blair, J. D., Hockemeyer, D., Doudna, J. A., Bateup, H. S. & Floor, S. N. Widespread translational remodeling during human neuronal differentiation. Cell Reports 21, 2005–2016 (2017).
Rodgers, J. T. et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature 510, 393–396 (2014).
Sahin, E. & DePinho, R. A. Axis of ageing: telomeres, p53 and mitochondria. Nat. Rev. Mol. Cell Biol. 13, 397–404 (2012).
Liu, H. K. et al. The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes Dev. 22, 2473–2478 (2008).
Werner, L. et al. Involvement of doublecortin-expressing cells in the arcuate nucleus in body weight regulation. Endocrinology 153, 2655–2664 (2012).
Shi, Z. et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83 (2017).
Anger, A. M. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013).
Retz, K. C. & Steele, W. J. Ribosome turnover in rat brain and liver. Life Sci. 27, 2601–2604 (1980).
Brannvall, K., Korhonen, L. & Lindholm, D. Estrogen-receptor-dependent regulation of neural stem cell proliferation and differentiation. Mol. Cell. Neurosci. 21, 512–520 (2002).
Rotheneichner, P. et al. Tamoxifen activation of Cre-recombinase has no persisting effects on adult neurogenesis or learning and anxiety. Front. Neurosci. 11, 27 (2017).
Mirzadeh, Z., Doetsch, F., Sawamoto, K., Wichterle, H. & Alvarez-Buylla, A. The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 39, e1938 (2010).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Dinkel, C., Moody, M., Traynor-Kaplan, A. & Schultz, C. Membrane-permeant 3-OH-phosphorylated phosphoinositide derivatives. Angew. Chem. Int. Edn Engl. 40, 3004–3008 (2001).
Lou, W. P., Baser, A., Klussmann, S. & Martin-Villalba, A. In vivo interrogation of central nervous system translatome by polyribosome fractionation. J. Vis. Exp. 30, e51255 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Acknowledgements
We thank S. Wolf from the DKFZ Genomics and Proteomics Core Facility; V. Eckstein from the Heidelberg University Hospital FACS Core Facility; M. Langlotz from the ZMBH FACS Core Facility; D. Wiest for the RPL22 antibody; K. Zwadlo, S. Limpert and K. Volk for technical assistance; D. Krunic and the Light Microscopy Core Facility for support with image analysis; B. Bukau and G. Kramer for advice during the early stage of the project, the TAC members A. Teleman, G. Stöcklin, M. Hentze and the members of the Martin-Villalba laboratory for critical comments and A. Teleman for critical reading of the manuscript. Y.D. was supported by China Scholarship Council (CSC). This work was supported by the University of Heidelberg and DKFZ (bridge-project ZMBH–DKFZ alliance), the DFG (SFB873; TRR186), the BMBF and the DKFZ. We are very grateful to our deceased friend, B. Fischer.
Reviewer information
Nature thanks S. McKnight, S.-B. Qian and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.M.-V. and A.B. conceived and designed the research. A.B., Y.D., M.S. and D.C.I. conducted studies on global protein synthesis, polysome profile and validation of transcriptome–translatome analysis. A.S.L. supported establishment of RiboTag models. G.S.G.B., C.S. and S.K. performed PtdIns(3,4,5)P3-related experiments. B.F., R.S. and M.G. performed computational analysis. E.Ll.B. and G.K. validated Cre-lines. A.M.-V., A.B. and B.F. wrote the manuscript. S.K. implemented all authors’ edited drafts. A.M.-V. and B.F. supervised, and A.M.-V. provided oversight of the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Validation of Tlx-eYFP reporter line, marker-based FACS isolation and protein synthesis in Tlx-lineage NSCs and ENBs.
a, FACS gates for NSCs labelled by TLX–tdTomato at four d.p.i. NSCs, which are labelled by recombination under the Tlx promoter, contain both quiescent (GLAST+EGFR−) and active NSCs (GLAST+EGFR+). PROM1 was not used in this experiment because TLX is not expressed in astrocytes. b, Composition of tdTomato+ cells four days after induction: 52.1 ± 0.6% are NSCs (GLAST+), 45.6% ± 0.3 are non-NSCs (GLAST−); they do not add up to 100% because a few cells were between gates. NSCs contain 51.7 ± 9.1% quiescent NSCs (EGFR−) and 46.7 ± 9.1% active NSCs (EGFR+). GLAST− cells contain 62.7 ± 1.6% neurogenic progenitors (NP; EGFR+PSA-NCAM−) and 33.3 ± 2% neuroblasts (EGFR−PSA-NCAM+). Results are presented from two biological replicates (n = 2); data are mean ± s.d. c, Scheme of the experimental design. Tlx-CreER mice were crossed with Rosa26-LSL-eYFP mice to generate Tlx-iYFP mice. Three groups of mice were treated with the indicated tamoxifen regimens by intraperitoneal injection and euthanized after two days. Each arrow represents 2 daily injections with a 12-h interval. d, e, Confocal image of the ventricular–subventricular zone (V-SVZ) of Tlx-iYFP mice (n = 5 mice) injected with tamoxifen as shown in c. d, Confocal image of the V-SVZ of a Tlx-iYFP mouse not injected with tamoxifen (top left panel) or injected with indicated doses of tamoxifen (other panels). Arrows mark ‘mislabelled’ parenchymal cells. Scale bars, 200 μm (main panels), 10 μm (insets). e, Immunostaining for the indicated markers. Arrowheads, GFAP-expressing eYFP-labelled NSCs; dashed line, ventricle surface. Scale bar, 50 μm. f, Schematic of tamoxifen administration, TLX-inducible-Cre-eYFP mouse model (TiCRY) and migrating path of eYFP NSCs lineages. g, Representative images of OPP incorporation of newly sorted eYFP+ NSCs and ENBs from the Tlx lineage were quantified. Scale bar, 10 µm. h, Quantification of OPP incorporation by NSCs (n = 38) and ENBs (n = 52) (relative to ENBs). Data are mean ± s.d. Two-tailed Student’s t-test; ****P < 0.0001. i, FACS strategy for isolation of quiescent and active NSCs and neuroblasts from wild-type mice.
Extended Data Fig. 2 Validation of specific recombination of RiboTag models in the targeted differentiation stages.
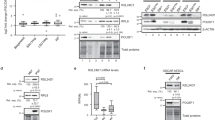
a, Top, NSCs isolated from TiCRY mice (n = 3) express HA tag. Replicates show different levels indicating variability based on induction efficiency. Cells from a wild-type mouse (n = 1) do not express the HA tag. Bottom, the HA tag is associated with RPL22, as staining shows a characteristic 8-kDa shift because of the HA tag (23 kDa instead of 15 kDa). Isolated TiCRY cells (n = 2 mice) entirely replaced wild-type RPL22 (WT) (n = 1 wild-type mouse) with RPL22–HA. Actin is shown as loading control. b, Representative Bioanalyzer profiles showing that input RNA levels do not vary between Cre+ TiCRY mice (n = 3) (expressing HA tag) and a Cre− TiCRY mouse (n = 1 mouse) (no HA tag). Samples from Cre+ mice reveal approximately threefold-more RNA over the background level in Cre− mice after HA immunoprecipitation. c, Enrichment of cell-type-specific transcripts by qRT–PCR after HA immunoprecipitation. Efficiency of enrichment of immunoprecipitation over input samples. Comparison of NSCs from NiCRY mice homozygous for the RPL22 tag (p/p; n = 2), heterozygous for the RPL22 tag (p/+; n = 2) and wild-type mice (+/+, n = 2) indicates a linear relationship between HA-tag expression and immunoprecipitation efficiency. NeuN serves as negative control. Cells isolated from NiCRY mice. Data are mean ± s.d. d, OPP incorporation does not vary between cells that express HA tag and cells that do not express HA tag, indicating no major effect on global protein synthesis. NSCs isolated from NiCRY mice. Scale bar, 10 µm. n = 30 cells per genotype. Data are mean ± s.d.; two-tailed Student’s t-test; P = 0.3739; n.s., not significant. e, Polysome fractionation using cultured NSCs indicates efficient incorporation of RPL22–HA (antibody against HA tag) into actively translating polysomes (see fractions 8–12). Total lysate resembles the positive control (+ctr). Cells from TiCRY mice. f, Disruption of polysomes using EDTA leads to shift of RPL22–HA expression to light fractions, indicating specific association with active ribosomes. e, f, Results are representative of n = 2 independent experiments. For gel source data (a, e, f), see Supplementary Fig. 1.
Extended Data Fig. 3 Validation of TiCRY and DiCRY mouse models.
a, Immunohistochemistry confirming HA-tag expression in NSCs of the SVZ at 4 d.p.i.; labelled cells are mostly GFAP+pH3−. eYFP+ cells are mostly NSCs (eYFP+DCX−), some ENBs (eYFP+DCX+) and a few of unknown identity (eYFP+DCX−, located in the striatum) (n = 3 mice). Scale bars, 50 µm. b, Immunohistochemistry confirming expression of HA tag in migrating neuroblasts, both in the SVZ–rostral migratory stream (RMS) (ENB, eYFP+DCX+) and the olfactory bulb (LNB, eYFP+DCX+NeuN−). Labelled cells within the SVZ of DiCRY mice are mostly ENBs, some of unknown identity (eYFP+DCX−) (n = 3 mice). Scale bars, 100 µm. c, Immunohistochemistry confirming expression of HA tag in NeuN+ neurons at 4 w.p.i. Cells were mostly mature NeuN+ neurons or NeuN− neurons in different layers of the olfactory bulb (n = 3 mice). Scale bar, 100 µm. d–g, Estimating cell-type composition based on eYFP expression and characteristic marker protein expression by immunofluorescence; see quantification in Fig. 2. Scale bars, 50 µm. d, NSCs in TiCRY mice (n = 3 mice) at 4 d.p.i. Green arrow, NSCs; yellow arrows, ENB contamination; white arrow, rare labelled cells outside the SVZ. e, LNBs in olfactory bulb of DiCRY mice (n = 3 mice) at 3 d.p.i.; labelled cells are mostly in the core of the olfactory bulb and express DCX protein. f, ENBs in SVZ of DiCRY mice (n = 3 mice) at 3 d.p.i. Yellow arrows, ENBs; white arrow, rare labelled cells outside the SVZ. g, Neurons in olfactory bulb of DiCRY mice (n = 3 mice) at 4 w.p.i.; labelled cells throughout the olfactory bulb mostly co-expressing NeuN protein. h, Time course of development of labelled cells in TiCRY mice. n = 1 mouse (3 d.p.i.), n = 3 mice (4 d.p.i.) and n = 2 mice ( 5 d.p.i.). Data are mean ± s.d. i, Schematic of experimental design; representative confocal images of the SVZ and olfactory bulb are taken at 3 d.p.i. or 4 w.p.i. (d–g). j, Note the presence of eYFP+ cells in the SVZ. Newly arriving eYFP+ cells are only present in the core of the olfactory bulb and absent from the other layers at 3 d.p.i.; after 4 w.p.i., the eYFP+ cells are absent from the SVZ, demonstrating that there is no residual Cre activity without tamoxifen induction. Furthermore, the core of the olfactory bulb does not contain eYFP+ cells, which are only present in the outer layers of the olfactory bulb where they have generated olfactory bulb interneurons. These experiments were performed in n = 3 biological replicates.
Extended Data Fig. 4 The RiboTag system allows parallel study of the transcriptome and translatome.
a, Enrichment of cell-type specific transcripts for NSC, ENB, LNB and neuron by fold enrichment of ribo-IP in Cre+ mice (n = 2) over Cre− mice (n = 2), assessed by qRT–PCR. Data are mean ± s.d. b, FACS strategy for isolation of NSCs and ENBs on the basis of eYFP expression. eYFP− cells were collected and used as control samples for enrichment analysis. c, Enrichment of cell-type specific transcripts by comparison of eYFP+ and eYFP− cells by qRT–PCR. All samples (n = 2 NSC; n = 3 ENB, LNB and neuron) were assessed by qRT–PCR before submission for sequencing. Data are mean ± s.d. d, Differential gene expression based on total mRNA. Subsequent populations were compared and significantly changing genes are shown in red (FDR = 10%). e, Differential gene expression based on ribo-IP sequencing without normalization for ribo-IP−. Subsequent populations were compared and significantly changing genes are marked in red (FDR = 10%). Data in d and e represent 2–3 biological replicates. Genes expected to be strongly represented after enrichment, corrected for each cell population, are labelled in red.
Extended Data Fig. 5 RiboTag system specifically targets each developmental stage along the neuronal differentiation lineage of adult olfactory bulb interneurons.
a, Expression of stage-specific marker genes for SVZ populations at the level of transcriptome (top) and translatome (bottom). b, Expression of stage-specific marker genes for olfactory bulb populations at the level of transcriptome (top) and translatome (bottom). Expression values based on transcripts per million (TPM) reads after trimmed mean of M values normalization. White bars in ribo-IP sequencing show background in ribo-IP− experiments (n = 2–3) (no HA tag expression), indicating average expression in the respective tissue corresponding to background binding (noise). In a and b, data are mean ± s.d. c–e, Gene set enrichment analysis on shared genes between total mRNA (n = 3) and ribo-IP sequencing (n = 2) for transition of NSC to ENB (c), ENB to LNB (d) and LNB to neuron (e, FDR = 10%) confirms enrichment of stage-specific gene ontologies. Only protein-coding genes are considered. f, Pearson's correlation coefficient was computed between the log2 read counts for different cell stages (NSC, ENB, LNB and neuron) of total mRNA sequencing (n = 3) and ribo-IP sequencing (n = 2) counts, showing agreement between the methods.
Extended Data Fig. 6 Assessment of translation efficiency using the RiboTag system.
a, Scheme describing the three datasets implemented into analysis of translation efficiency. b, Summary of the fraction of genes repressed (NSC: 32/4,500; ENB: 282/3,445; LNB: 622/1,958; neurons: 714/1,443; FDR = 10%) or enhanced (NSC: 9/4,500; ENB: 63/3,445; LNB: 225/1,958; neurons: 214/1,443; FDR = 10%) in each population. c, Left, summary of the absolute number of genes that are repressed or enhanced in each population (protein-coding genes only). Remaining panels show the comparison of repressed and enhanced genes at different stages, revealing increasing overlap with progression of cells. LNBs and neurons feature high overlap of repressed genes (n = 311). d, Analysis of explained variance scoring the contribution of background (ribo-IP−), total mRNA, translation efficiency (ribo-IP+ sequencing over total mRNA and ribo-IP−) and unknown residual noise to the ribo-IP+ sequencing data of each population. e, As in d, but comparing only the proportion of variance explained by translation efficiency and total mRNA. Translation efficiency explains more of the data in neurons compared to earlier developmental stages and NSCs, indicating increased importance of translational regulation in neurons. Analysis in d and e was performed from three biological replicates (n = 3). f, Assessment of mRNA levels by qRT–PCR of several candidate genes in freshly isolated NSCs and ENBs normalized to Actb (left). Pearson's correlation coefficient computed between the log2 read counts from NSCs to ENBs of total mRNA (right) counts and qRT-PCR values. This was done to demonstrate the agreement between the methods. g, Left, representative western blot showing protein expression of candidate genes in freshly sorted NSCs and ENBs (n = 15 pooled mice). Vinculin is shown on the same membrane, as loading control. Right, Pearson's correlation coefficient computed between the log2 read counts from NSCs to ENBs of the translation efficiency estimated from high-throughput sequencing and the intensities from western blots. This was done to demonstrate the agreement between the methods. For gel source data, see Supplementary Fig. 1.
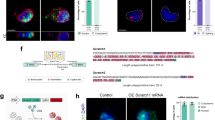
Extended Data Fig. 7 Parallel in situ hybridization and immunohistochemistry for detection of Sox2 mRNA and protein expression along the neuronal lineage.
a, Quantification of Sox2 RNA and SOX2 protein. The regression lines indicate different translation efficiencies. b, Representative images for parallel in situ hybridization and immunocytochemistry of freshly sorted cells of the SVZ. Comparison of NSCs (GLAST+PROM+), neurogenic progenitors (NPs, GLAST−PROM−EGFR+) and ENBs (GLAST−PROM−PSA-NCAM+) stained for cell-type-specific marker proteins and SOX2 protein. In situ hybridization for Sox2 mRNA. Scale bar, 5 µm. Data are representative of two independent experiments. c, Quantification of SOX2 protein and RNA expression in NSCs (n = 22), neurogenic progenitors (n = 49) and ENBs (n = 35). Regression lines indicate that translation is most efficient in NSCs. d, Ratio of protein expression to RNA abundance indicates higher translation efficiency of SOX2 in NSCs when compared to neurogenic progenitors or ENBs. Same replicates were used as in c. Data are mean ± s.d.; two-tailed Student’s t-test; *P = 0.0279, *P = 0.021. e, Representative image for parallel in situ hybridization for Sox2 mRNA in NSCs (marked by TLX-tdTomato) and ENBs (marked by DCX). Parallel detection of DapB (bacterial transcript) and Ppib (ubiquitous transcript) served as negative and positive controls, respectively. Scale bar, 25 µm. Data are representative for two independent experiments (n = 2). f, SOX2 expression in calretinin+DCX–eYFP+ cells (4 w.p.i.) in the PGL of the olfactory bulb (n = 1 mouse). Scale bar, 50 µm. g, Detection of SOX2 protein in short-term (3 d.p.i.) and long-term (4 w.p.i.) traced DCX–eYFP+ cells, indicating the contribution of migrating, newly differentiated neurons to the pool of SOX2+ neurons in the periglomerular layer. Scale bar, 50 µm. n = 5 mice, 3 d.p.i.; n = 4 mice, 4 w.p.i.; two-tailed Mann–Whitney U-test; *P = 0.0159. Data are mean ± s.d.
Extended Data Fig. 8 A drop in mTORC1 activity is required for stem cell exit and onset of neuronal differentiation.
a, Gene Ontology analysis for genes either repressed (top) or enhanced (bottom) at the ENB stage. The repressed genes are mostly related to the protein synthesis machinery, whereas many upregulated genes are involved in development, differentiation and neuronal function. Gene Ontology is based on three biological replicates. b, Stage-specific repression of 5′-PRM-containing transcripts in ENBs and not LNBs, independent of the total number of repressed genes (1% FDR). c, Staining for pS6 in the SVZ as readout for mTORC1 activity. Comparison of NSCs (TLX–eYFP+) and ENBs (DCX+). All ENBs show low levels of pS6 (green arrow), whereas some NSCs show high pS6 (orange arrows) and others show low levels of pS6 (yellow arrow). Scale bar, 50 µm. Images are representative of two biological replicates. d, Western blot analysis of the steady-state level of mTOR signalling components in NSCs and ENBs freshly sorted from the SVZ. Results are representative of n = 2 independent experiments. Vinculin is shown on the same membrane as loading control. For gel source data, see Supplementary Fig. 1. e, Staining for pS6 in the olfactory bulb as readout for mTORC1 activity. The core of the olfactory bulb (marked with dashed line) which contains only LNBs (DCX+) is enriched for high pS6, whereas only a few neurons (NeuN+DCX−) have high levels of pS6. Scale bar, 50 µm. Images are representative of three biological replicates. f, Representative absorption profiles for DMSO and Torin-treated NSCs (2 h) after sucrose gradient fractionation indicate slight repression in global translation upon Torin treatment. Representative plots showing the distribution of mRNAs for some genes across the gradient fractions of the polysome profiles are shown. Note a strong shift of PRM+ mRNAs (Rpl18, eIF3f, eEF1b2, Rps17) but not PRM− mRNAs (Actb, Sox9) to the fractions of light polysomes and non-translating complexes following Torin treatment. Sox2 transcript distribution across the polysome fraction was not influenced following 2 h Torin treatment. Data are mean ± s.d.; three biological replicates. g, Representative images of SOX2 and DCX expression in NSCs exposed to DMSO (Co), PtdIns(3,4,5)P3 or PtdIns(3,4,5)P3 + Torin after 7 days in vitro culture under differentiating conditions. Scale bar, 10 µm. h, Representative images of SOX2 and pS6 expression of NSCs and freshly isolated ENBs exposed to DMSO (Co), PtdIns(3,4,5)P3, or PtdIns(3,4,5)P3 + Torin for 2 h. Scale bar, 5 µm.
Extended Data Fig. 9 Orchestration of mTORC1 and cell cycle enables post-transcriptional repression of SOX2 expression.
a, Western blot and corresponding quantification of vehicle or Torin-treated primary NSCs showing inhibition of p70 S6K and rpS6 phosphorylation, as well as 4E-BP phosphorylation, all indicators of mTORC1 activity. mTORC1 regulators TSC2 and RHEB, as well as SOX2, are not affected by 2 h incubation with Torin. b, Post-transcriptional repression by Torin was assessed in synchronized primary NSCs (passage 2) at different time points by western blot. Corresponding quantification of protein levels for SOX2 and phosphorylation of p70 S6K and 4E-BP is shown (right). c, Western blot of primary NSCs treated for 4 h with erlotinib reveals a drop in EGFR phosphorylation accompanied by very low levels of phosphorylated p70 S6K and pS6. TSC2, RHEB and SOX2, as with Torin treatment, remain at a constant level. d, Genes translationally upregulated upon the transition from NSCs to ENBs in vivo show slightly higher protein abundance following 2-h treatment with Torin in unsynchronized NSCs. e, In synchronized NSCs, some of these genes show higher protein abundance either through cell-cycle exit and/or Torin treatment. In a–e, western blots are representative of two independent experiments. Vinculin is shown on the same membrane as loading control. For gel source data, see Supplementary Fig. 1. f, Graphical abstract summarizing the findings of this study. Using the RiboTag system for parallel assessment of transcriptome and translatome along the neuronal differentiation lineage reveals post-transcriptional regulation of stem cell exit by mTORC1 at late G1 state.
Supplementary information
Supplementary Information
This file contains an Index for Supplementary Information.
Supplementary Figures
This file contains Supplementary Figure 1: uncropped gel scans.
Supplementary Table
This file contains Supplementary Table S1 Normalized Reads for Single Samples. Data are presented from n=3 biological replicates. Adjusted p-value; adjusted by the method of Benjamini-Hochberg controlling for FDR. Significance code: * = 10% FDR; ** = 1% FDR.
Supplementary Table
This file contains Supplementary Table S2 Differential Expression RNAseq. Data are presented from n=3 biological replicates. Adjusted p-value; adjusted by the method of Benjamini-Hochberg controlling for FDR. Significance code: * = 10% FDR; ** = 1% FDR.
Supplementary Table
This file contains Supplementary Table S3 Differential Expression RIBOseq. Data are presented from n=2 biological replicates. Adjusted p-value; adjusted by the method of Benjamini-Hochberg controlling for FDR. Significance code: * = 10% FDR; ** = 1% FDR.
Supplementary Table
This file contains Supplementary Table S4 Translation Efficiency. Data are presented from n=3 biological replicates. Adjusted p-value; adjusted by the method of Benjamini-Hochberg controlling for FDR. Significance code: * = 10% FDR; ** = 1% FDR.
Supplementary Table
This file contains Supplementary Table S5 unbiased motif analysis. Data are presented from n=3 biological replicates. Adjusted p-value; adjusted by the method of Benjamini-Hochberg controlling for FDR. Significance code: * = 10% FDR; ** = 1% FDR.
Source data
Rights and permissions
About this article
Cite this article
Baser, A., Skabkin, M., Kleber, S. et al. Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100–104 (2019). https://doi.org/10.1038/s41586-019-0888-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-0888-x
This article is cited by
-
Astrocytes in the adult dentate gyrus—balance between adult and developmental tasks
Molecular Psychiatry (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
Intron detention tightly regulates the stemness/differentiation switch in the adult neurogenic niche
Nature Communications (2024)
-
Regulation of adult stem cell quiescence and its functions in the maintenance of tissue integrity
Nature Reviews Molecular Cell Biology (2023)
-
Effects of low doses of methylmercury (MeHg) exposure on definitive endoderm cell differentiation in human embryonic stem cells
Archives of Toxicology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.