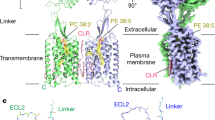
Abstract
Type-A γ-aminobutyric (GABAA) receptors are ligand-gated chloride channels with a very rich pharmacology. Some of their modulators, including benzodiazepines and general anaesthetics, are among the most successful drugs in clinical use and are common substances of abuse. Without reliable structural data, the mechanistic basis for the pharmacological modulation of GABAA receptors remains largely unknown. Here we report several high-resolution cryo-electron microscopy structures in which the full-length human α1β3γ2L GABAA receptor in lipid nanodiscs is bound to the channel-blocker picrotoxin, the competitive antagonist bicuculline, the agonist GABA (γ-aminobutyric acid), and the classical benzodiazepines alprazolam and diazepam. We describe the binding modes and mechanistic effects of these ligands, the closed and desensitized states of the GABAA receptor gating cycle, and the basis for allosteric coupling between the extracellular, agonist-binding region and the transmembrane, pore-forming region. This work provides a structural framework in which to integrate previous physiology and pharmacology research and a rational basis for the development of GABAA receptor modulators.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available from the corresponding authors and/or included in the manuscript or Supplementary Information. Atomic coordinates for PTX-, PTX/GABA-, BCC-, ALP/GABA- and DZP/GABA-bound structures have been deposited in the Protein Data Bank with accession codes 6HUG, 6HUJ, 6HUK, 6HUO and 6HUP, respectively, and the cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-0275, EMD-0279, EMD-0280, EMD-0282 and EMD-0283, respectively.
Change history
07 February 2019
In Fig. 5b, d, the arrows showing transmembrane domain rotations were inadvertently pointing clockwise instead of anticlockwise. Similarly, ‘anticlockwise’ should have been ‘clockwise’ in the sentence ‘This conformational change of the ECD triggers a clockwise rotation of the TMD.’ In Extended Data Table 1, the units of the column ‘Model resolution’ should have been Å instead of Å2. These errors have been corrected online.
References
Sieghart, W. Structure and pharmacology of γ-aminobutyric acidA receptor subtypes. Pharmacol. Rev. 47, 181–234 (1995).
Barnard, E. A. et al. International Union of Pharmacology. XV. Subtypes of γ-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol. Rev. 50, 291–313 (1998).
Sigel, E. & Steinmann, M. E. Structure, function, and modulation of GABAA receptors. J. Biol. Chem. 287, 40224–40231 (2012).
Möhler, H. GABAA receptors in central nervous system disease: anxiety, epilepsy, and insomnia. J. Recept. Signal Transduct. 26, 731–740 (2006).
Olsen, R. W. Analysis of γ-aminobutyric acid (GABA) type A receptor subtypes using isosteric and allosteric ligands. Neurochem. Res. 39, 1924–1941 (2014).
Olsen, R. W. Allosteric ligands and their binding sites define γ-aminobutyric acid (GABA) type A receptor subtypes. Adv. Pharmacol. 73, 167–202 (2015).
Hunkeler, W. Benzodiazepines, the story of the antagonist flumazenil and of the partial agonist bretazenil. Chimia 47, 141–147 (1993).
Squires, R. F. & Brastrup, C. Benzodiazepine receptors in rat brain. Nature 266, 732–734 (1977).
Sigel, E., Mamalaki, C. & Barnard, E. A. Isolation of a GABA receptor from bovine brain using a benzodiazepine affinity column. FEBS Lett. 147, 45–48 (1982).
Schofield, P. R. et al. Sequence and functional expression of the GABAA receptor shows a ligand-gated receptor super-family. Nature 328, 221–227 (1987).
Hibbs, R. E. & Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011).
Miyazawa, A., Fujiyoshi, Y. & Unwin, N. Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955 (2003).
Miller, P. S. & Aricescu, A. R. Crystal structure of a human GABAA receptor. Nature 512, 270–275 (2014).
Miller, P. S. et al. Structural basis for GABAA receptor potentiation by neurosteroids. Nat. Struct. Mol. Biol. 24, 986–992 (2017).
Laverty, D. et al. Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites. Nat. Struct. Mol. Biol. 24, 977–985 (2017).
Chen, Q. et al. Structural basis of neurosteroid anesthetic action on GABAA receptors. Nat. Commun. 9, 3972 (2018).
Liu, S. et al. Cryo-EM structure of the human α5β3 GABAA receptor. Cell Res. 28, 958–961 (2018).
Zhu, S. et al. Structure of a human synaptic GABAA receptor. Nature 559, 67–72 (2018).
Phulera, S. et al. Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. eLife 7, e39383 (2018).
Miller, P. et al. Heteromeric GABAA receptor structures in positively-modulated active states. Preprint at https://www.biorxiv.org/content/early/2018/06/05/338343 (2018).
Baumann, S. W., Baur, R. & Sigel, E. Individual properties of the two functional agonist sites in GABAA receptors. J. Neurosci. 23, 11158–11166 (2003).
Tan, K. R., Rudolph, U. & Lüscher, C. Hooked on benzodiazepines: GABAA receptor subtypes and addiction. Trends Neurosci. 34, 188–197 (2011).
Knoflach, F. et al. Pharmacological modulation of the diazepam-insensitive recombinant gamma-aminobutyric acidA receptors alpha 4 beta 2 gamma 2 and alpha 6 beta 2 gamma 2. Mol. Pharmacol. 50, 1253–1261 (1996).
Johnston, G. A. R. Advantages of an antagonist: bicuculline and other GABA antagonists. Br. J. Pharmacol. 169, 328–336 (2013).
Krishek, B. J., Moss, S. J. & Smart, T. G. A functional comparison of the antagonists bicuculline and picrotoxin at recombinant GABAA receptors. Neuropharmacology 35, 1289–1298 (1996).
Goutman, J. D. & Calvo, D. J. Studies on the mechanisms of action of picrotoxin, quercetin and pregnanolone at the GABAρ 1 receptor. Br. J. Pharmacol. 141, 717–727 (2004).
Xu, X.-J., Roberts, D., Zhu, G.-N. & Chang, Y.-C. Competitive antagonists facilitate the recovery from desensitization of α1β2γ2 GABAA receptors expressed in Xenopus oocytes. Acta Pharmacol. Sin. 37, 1020–1030 (2016).
Dostalova, Z. et al. Human α1β3γ2L gamma-aminobutyric acid type A receptors: high-level production and purification in a functional state. Protein Sci. 23, 157–166 (2014).
Laverty, D. et al. Cryo-EM structure of the human α1β3γ2 GABAA receptor in a lipid bilayer. Nature https://doi.org/10.1038/s41586-018-0833-4 (2019).
Chen, L., Durkin, K. A. & Casida, J. E. Structural model for gamma-aminobutyric acid receptor noncompetitive antagonist binding: widely diverse structures fit the same site. Proc. Natl Acad. Sci. USA 103, 5185–5190 (2006).
Xu, M., Covey, D. F. & Akabas, M. H. Interaction of picrotoxin with GABAA receptor channel-lining residues probed in cysteine mutants. Biophys. J. 69, 1858–1867 (1995).
Gurley, D., Amin, J., Ross, P. C., Weiss, D. S. & White, G. Point mutations in the M2 region of the alpha, beta, or gamma subunit of the GABAA channel that abolish block by picrotoxin. Recept. Channels 3, 13–20 (1995).
Bali, M. & Akabas, M. H. The location of a closed channel gate in the GABAA receptor channel. J. Gen. Physiol. 129, 145–159 (2007).
Wang, D.-S., Mangin, J.-M., Moonen, G., Rigo, J.-M. & Legendre, P. Mechanisms for picrotoxin block of α2homomeric glycine receptors. J. Biol. Chem. 281, 3841–3855 (2006).
Unwin, N. Nicotinic acetylcholine receptor and the structural basis of neuromuscular transmission: insights from Torpedo postsynaptic membranes. Q. Rev. Biophys. 46, 283–322 (2013).
Qian, H., Pan, Y., Zhu, Y. & Khalili, P. Picrotoxin accelerates relaxation of GABAC receptors. Mol. Pharmacol. 67, 470–479 (2005).
Chang, Y. & Weiss, D. S. Site-specific fluorescence reveals distinct structural changes with GABA receptor activation and antagonism. Nat. Neurosci. 5, 1163–1168 (2002).
Mehta, A. K. & Ticku, M. K. Characterization of the picrotoxin site of GABAA receptors. Curr. Protoc. Pharmacol. 8, (2001).
Talwar, S. & Lynch, J. W. Phosphorylation mediated structural and functional changes in pentameric ligand-gated ion channels: implications for drug discovery. Int. J. Biochem. Cell Biol. 53, 218–223 (2014).
Olsen, R. W. GABAA receptor: Positive and negative allosteric modulators. Neuropharmacology 136 (Pt A), 10–22 (2018).
Chang, Y., Ghansah, E., Chen, Y., Ye, J. & Weiss, D. S. Desensitization mechanism of GABA receptors revealed by single oocyte binding and receptor function. J. Neurosci. 22, 7982–7990 (2002).
Gielen, M. & Corringer, P.-J. The dual-gate model for pentameric ligand-gated ion channels activation and desensitization. J. Physiol. (Lond.) 596, 1873–1902 (2018).
Ueno, S., Bracamontes, J., Zorumski, C., Weiss, D. S. & Steinbach, J. H. Bicuculline and gabazine are allosteric inhibitors of channel opening of the GABAA receptor. J. Neurosci. 17, 625–634 (1997).
Middendorp, S. J. et al. Relative positioning of classical benzodiazepines to the γ2-subunit of GABAA receptors. ACS Chem. Biol. 9, 1846–1853 (2014).
Wieland, H. A., Lüddens, H. & Seeburg, P. H. A single histidine in GABAA receptors is essential for benzodiazepine agonist binding. J. Biol. Chem. 267, 1426–1429 (1992).
Rudolph, U. & Knoflach, F. Beyond classical benzodiazepines: novel therapeutic potential of GABAA receptor subtypes. Nat. Rev. Drug Discov. 10, 685–697 (2011).
Forman, S. A. & Miller, K. W. Anesthetic sites and allosteric mechanisms of action on Cys-loop ligand-gated ion channels. Can. J. Anaesth. 58, 191–205 (2011).
Walters, R. J., Hadley, S. H., Morris, K. D. W. & Amin, J. Benzodiazepines act on GABAA receptors via two distinct and separable mechanisms. Nat. Neurosci. 3, 1274–1281 (2000).
Fisher, J. L. A lysine residue in the β3 subunit contributes to the regulation of GABAA receptor activity by voltage. Mol. Cell. Neurosci. 20, 683–694 (2002).
Renaud, J.-P. et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 (2018).
Einhauer, A. & Jungbauer, A. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem. Biophys. Methods 49, 455–465 (2001).
Molday, R. S. & MacKenzie, D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22, 653–660 (1983).
Ritchie, T. K. et al. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009).
Javaheri, A. et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2, 16189 (2016).
Pardon, E. et al. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ https://doi.org/10.1107/S205225251801463X (2019).
Vilas, J. L. et al. MonoRes: automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D 74, 814–840 (2018).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996).
Forman, S. A. A hydrophobic photolabel inhibits nicotinic acetylcholine receptors via open-channel block following a slow step. Biochemistry 38, 14559–14564 (1999).
Acknowledgements
We thank G. Cannone and S. Chen for cryo-EM assistance at the MRC-LMB; Y. Chaban and K. Dent for cryo-EM assistance at the Electron Bio-Imaging Centre (eBIC), Diamond Light Source; J. García-Nafría, L. Dong, T. Nakane and S. Scheres for advice on cryo-EM data processing; J. Grimmett and T. Darling for IT and computing support; and members of the Aricescu laboratory for discussions and comments on the manuscript. This work was supported by the UK Medical Research Council grants MR/L009609/1, MC_UP_1201/15 (A.R.A., S.M. and D.L.) and MC_UP_A025_1013 (J.Z.); UK Biotechnology and Biological Sciences Research Council grant BB/M024709/1 (A.R.A. and D.L.); Human Frontier Science Program grant RGP0065/2014 (A.R.A.); Cancer Research UK grant C20724/A14414 (T.M.); and Swiss National Science Foundation fellowship 168735 (J.Z.). R.D. and K.W.M. were supported by a grant from the National Institute for General Medical Sciences (GM 58448) and by the Department of Anesthesia, Critical Care and Pain Medicine, Massachusetts General Hospital. We acknowledge the support and the use of resources of Instruct-ERIC (PID1271), part of the European Strategy Forum on Research Infrastructures (ESFRI), and the Research Foundation-Flanders (FWO) for their support of nanobody discovery, and FWO for a doctoral fellowship to T.U.
Reviewer information
Nature thanks M. Chebib, M. Jansen and H. Nury for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
S.M. and A.R.A. conceived the project. S.M. carried out protein purification, collected and processed the cryo-EM data, and built and refined the models, with assistance from A.R.A. R.D. and K.W.M. designed and analysed the electrophysiological experiments, which were performed by R.D. T.U., E.P. and J.S. designed and generated Mb38. I.S.M. contributed to sample screening by cryo-EM. D.L. developed Volta Phase Plate data collection protocols and contributed to model building. D.K. and A.K. contributed to cryo-EM data collection on the Krios-S microscope. T.M. contributed to the analysis of structural data. J.Z. developed contrast transfer function refinement algorithms. S.M. and A.R.A. wrote the manuscript with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Single-particle cryo-EM analysis of human α1β3γ2L GABAA receptor bound to the channel-blocker picrotoxin.
a, Representative micrograph of the PTX-bound GABAA receptor particles embedded in vitreous ice. b, Representative 2D class averages. c, FSC curves for the 3D reconstruction using gold-standard refinement in RELION56. Curves are shown for the phase randomization, unmasked, masked and phase-randomization-corrected masked maps. d, Validation of model refinement protocol. Curves are shown for model versus summed map (FSCfull), model refined in half-map 1 versus half-map 1 (FSCwork) and model refined in half-map 1 versus half-map 2 (FSCfree). e, The final, unsharpened cryo-EM map coloured by local resolution (estimated using MonoRes61) shown at a higher contour level (left) and at a lower level (middle and right) to highlight the nanodisc belt and flexible intracellular domains (ICDs). f, Angular distribution of particle projections. The map of the GABAA receptor–PTX complex is shown in teal. g, Cryo-EM density segments for the PTX-binding site between residues 2ʹ and 9ʹ of the M2 transmembrane helices.
Extended Data 2 Structural and electrophysiological analyses of human α1β3γ2L GABAA receptor in complex with PTX and GABA.
a, FSC curves for the 3D reconstruction of the GABAA receptor bound to PTX and GABA. Curves are shown for the phase randomization, unmasked, masked and phase-randomization-corrected masked maps. b, Validation of the model refinement protocol. Curves are shown for model versus summed map (FSCfull), model refined in half-map 1 versus half-map 1 (FSCwork) and model refined in half-map 1 versus half-map 2 (FSCfree). c, The final, unsharpened cryo-EM map coloured by local resolution (estimated using MonoRes61). d, Superposition of the PTX-bound and PTX/GABA-bound α1β3γ2 receptor based on the global TMD alignment. The GABA-induced movements of loop-C in each of the β3 subunits are highlighted by green lines between Cα atoms of Thr202 residues. GABA is shown as spheres (carbon, khaki; nitrogen, blue; oxygen, red). e, Cryo-EM density segments showing GABA-binding sites in the PTX/GABA-bound structure. f–h, Representative whole-cell current traces elicited from the same HEK293 cell by three 8.8-s pulses of GABA (5 μM) plus Mb38 (2 μM), each separated by a 1-min wash: control (f); one second after the start of the second 8.8-s pulse, PTX (800 μM) was co-applied for 4 s (g); wash control showing full recovery (h). PTX inhibited currents by 106 ± 2.6% (mean ± s.d.; n = 6 cells). In addition, the protocol was repeated with outside-out patches (117 ± 9% (mean ± s.d.); n = 5 patches). i, Globally superposed PTX-bound and PTX/GABA-bound α1β3γ2 receptor transmembrane domains viewed from the extracellular space. Side chains of 9ʹ Leu residues are shown as sticks, whereas PTX is represented as balls and sticks. j, k, Superposition of α1 subunit ECDs from PTX-bound and PTX/GABA-bound α1β3γ2 receptors reveal the relative β3 ECD motions towards α1 ECDs, as viewed from outside of the receptor (j) and from the vestibule (k). Differences in distances (Å) between the selected Cα atoms in the complexes without and with GABA are indicated by lines. The PTX-bound structure is shown in grey and the PTX/GABA-bound structure is coloured by subunit (α1, red; β3, blue; γ2, yellow).
Extended Data Fig. 3 Structural analysis of human α1β3γ2L GABAA receptor bound to the competitive antagonist bicuculline.
a, FSC curves for the 3D reconstruction of the GABAA receptor bound to BCC. Curves are shown for the phase randomization, unmasked, masked and phase-randomization-corrected masked maps. b, Validation of the model refinement protocol. Curves are shown for model versus summed map (FSCfull), model refined in half-map 1 versus half-map 1 (FSCwork) and model refined in half-map 1 versus half-map 2 (FSCfree). c, The final, unsharpened cryo-EM map coloured by local resolution (estimated using MonoRes61). d, Cryo-EM density maps of the BCC-binding pockets. e, Superposition of the PTX-bound and BCC-bound α1β3γ2 receptor based on the global TMD alignment. The BCC-induced movements of loop-C in each of the β3 subunits are highlighted by green lines between Cα atoms of Thr202 residues. BCC is shown as spheres (carbon, khaki; nitrogen, blue; oxygen, red). f, Superposition of individual subunits from the PTX-bound and BCC-bound GABAA receptor structures. r.m.s.d. values (Å) for equivalent Cα in the entire subunits are shown. Loops-C are marked by arrows. The PTX-bound structure is shown in grey and the BCC-bound structure is coloured by subunit (α1, red; β3, blue; γ2, yellow).
Extended Data Fig. 4 Structural analysis of human α1β3γ2L GABAA receptor in complexes with alprazolam and diazepam.
a, FSC curves for the 3D reconstruction of the GABAA receptor bound to ALP. Curves are shown for the phase randomization, unmasked, masked and phase-randomization-corrected masked maps. b, Validation of the model refinement protocol. Curves are shown for model versus summed map (FSCfull), model refined in half-map 1 versus half-map 1 (FSCwork) and model refined in half-map 1 versus half-map 2 (FSCfree). c, The final, unsharpened cryo-EM map coloured by local resolution (estimated using MonoRes61). d–f, Same as a–c but for the GABAA receptor bound to DZP. g–i, Cryo-EM density maps of the ligand binding sites: ALP binding in the BZD pocket (g), DZP binding in the BZD pocket (h), DZP binding in the general-anaesthetic pocket at the β3+/α1− interface (i). Ligands are shown as sticks, with atom colouring as follows: ALP carbons, cyan; DZP carbons, teal; nitrogen, blue; chlorine, green. Side chains of residues lining the binding pockets are shown as sticks and are numbered. Dotted circles highlight the difference between the structures of alprazolam and diazepam.
Extended Data Fig. 5 The classical benzodiazepines flumazenil and bretazenil bind to the same benzodiazepine pocket in GABAA receptors, but use different modes.
ALP/GABA-bound and DZP/GABA-bound structures are coloured by subunit (α1, red; β3, blue; γ2, yellow), whereas the other superposed structures are shown in grey. Loop-C is shown in a coil representation, to enable better visualization of the BZD pocket. a, Structural formulae of flumazenil (FZL) and (S)-bretazenil (BRZ). b, c, Superposition of γ2 subunit ECDs from the DZP-bound α1β3γ2 and FZL-bound α1β2γ2 receptor structures reveals the FZL16 (white) position in the BZD pocket relative to DZP (teal). Side-on (b) and top-down (c) views of the pocket are presented. d, e, Same as b, c but the structural alignment shows the relative position of the BRZ15 (white). Grey dashed lines indicate hydrogen bonds that FZL and BRZ form with γ2Thr142 in the BZD pocket. f, Superposition of γ2 subunit ECDs from the PTX-, PTX/GABA-, BCC-, DZP/GABA- to ALP/GABA-bound γ2 ECD illustrates conformational changes of the BZD pocket associated with ALP/DZP binding: an outward movement of loop-C; rearrangement of γ2Tyr58 and γ2Phe77 side chains; and a change in the γ2Asn60 rotamer. g, Superposition of the α1-D subunit ECDs from the PTX-bound and ALP/GABA-bound α1β3γ2 structures shows that ALP binding causes only a minimal outwards motion of the α1 loop-C, by 0.8 Å as measured between Ser206 Cα atom positions.
Extended Data Fig. 6 Superposition of individual subunits from ALP-bound and DZP-bound α1β3γ2L GABAA receptor structures.
Superposition of the individual subunits from the DZP-bound (grey) and ALP-bound (α1, red; β3, blue; γ2, yellow) GABAA receptor structures. r.m.s.d. values are in the range of 0.38–0.41 Å for α1 (343–344 equivalent Cα positions), 0.37–0.40 Å for β3 (334–336 equivalent Cα positions) and 0.47 Å for γ2 subunits (330 equivalent Cα positions). Loops-C are marked by arrows.
Extended Data Fig. 7 Structural analysis of PTX-bound and ALP/GABA-bound α1β3γ2L GABAA receptor structures.
The PTX-bound structure is shown in grey and the ALP/GABA-bound structure is coloured by subunit (α1, red; β3, blue; γ2, yellow). a, b, Superposition of α1 subunit ECDs from PTX-bound and ALP/GABA-bound GABAA receptors reveals the relative β3 ECD motions towards α1 ECDs, as viewed from outside of the receptor (a) and from the vestibule (b). Differences in distances (Å) between the selected Cα atoms in the complexes without and with GABA are indicated with lines. c, Individual subunits from the PTX-bound and the ALP/GABA-bound GABAA receptor structures superposed on the basis of the global TMD alignment (ALP/GABA TMD over PTX TMD). Angles between vectors representing M2 helices and the pore axis of the PTX-bound structure are shown. Side chains of residues at the −2ʹ and the 9ʹ positions are shown. d, Superposition of TMDs from PTX-bound and ALP/GABA-bound structures. r.m.s.d. values (Å) are shown for entire TMDs and for the M2–M3 loops (see Methods for boundary definitions).
Extended Data Fig. 8 Conformational differences at the ECD–TMD interfaces between PTX-bound (closed) and ALP/GABA-bound (desensitized) α1β3γ2L GABAA receptor structures.
The PTX-bound structure is shown in grey and the ALP/GABA-bound structure is coloured by subunit (α1, red; β3, blue; γ2, yellow). The TMDs of the principal subunits of PTX-bound and ALP/GABA-bound structures were superposed enabling visualization of relative movements of neighbouring ECDs and TMDs. a, b, Structural rearrangements of the ECD–TMD interface between α1-A and β3-E subunits. c, d, Same as a, b, but for γ2-C and β3-B subunits. e, f, Same as a, b, but for β3-B and α1-A subunits. Amino acid residues present in the β1–β2 loop tip in each subunit are shown, the Cαs for these residues are represented as spheres and the distances of displacement indicated. The strictly conserved M2–M3 loop proline residue interacting with the β1–β2 loop is shown for each subunit. β1–β2 loop motions are indicated by curved arrows. g–l, Conformational differences in the kM2–M3 loop and 19ʹ Arg side chain positions between the PTX-bound and the ALP/GABA-bound structures shown for α-A (g, h), g2-C (i, j) and b3-B (k, l) subunits. Neighbouring subunit M1 and M2 helices are shown as cylinders. Dashed lines indicate putative hydrogen bond interactions between amino acid side chains and main-chain carbonyls.
Supplementary information
Video 1
Structure of the human α1β3γ2L GABAAR in complex with picrotoxin. The video starts with a 360° rotation of the GABAAR cryo-EM map (α1, red; β3, blue; γ2, yellow; glycan, orange) in complex with Mb38 (green), phosphatidylinositol 4,5-bisphosphate (PIP2, light brown) and picrotoxin (PTX, pink) parallel to the plane of the membrane. Next, the cryo-EM density is sliced from top to bottom, revealing empty orthosteric sites (indicated with dashed grey wedges) and the PTX bound to the channel pore, between the 9' and 2' amino acid rings. The PTX binding site is also shown form a view parallel to the membrane.
Video 2
Picrotoxin binds within the transmembrane pore of the human α1β3γ2L GABAAR. Architecture of the PTX binding site. PTX is shown (ball-and-stick depiction, carbon atoms are in pink; oxygen, red) bound in between the 9' Leu ring (proximal to the extracellular region) and the 2' ring (Val257 in α1; Ala252 in β3; Ser267 in γ2) (proximal to the cytoplasmic region). Side chains of residues proximal to PTX are shown as sticks and are numbered (oxygen, red; colouring of carbon atoms matches the corresponding subunit colouring). The PTX cryo-EM density is shown as a grey surface.
Video 3
Structure of the human α1β3γ2L GABAAR in complex with picrotoxin and GABA. The video first shows a 360° rotation of the GABAAR cryo-EM map (α1, red; β3, blue; γ2, yellow; glycan, orange) in complex with Mb38 (green), PIP2 (light brown), PTX (pink) and GABA (khaki) parallel to the plane of the membrane. Then, the cryo-EM density is shown gradually sliced from top to bottom, revealing orthosteric binding sites occupied with GABA and the transmembrane domain bound to PTX.
Video 4
γ-aminobutyric acid (GABA) binding sites in the human α1β3γ2L GABAAR. Close-up of the GABA binding pocket at the β3-E+/α1-D− interface in the PTX/GABA-bound α1β3γ2L receptor structure. GABA is shown as sticks (carbon atoms are in khaki; nitrogen, blue; oxygen, red). Side chains of residues lining the pocket are shown as sticks (nitrogen, blue; oxygen, red) and numbered. The cryo-EM density of GABA is shown as a grey surface.
Video 5
Conformational differences between picrotoxin- (closed/resting) and picrotoxin/GABA-bound (pre-active) human α1β3γ2L GABAAR structures. This video shows a morph between PTX- and PTX/GABA-bound structures aligned using their transmembrane domains (TMDs). The morph is sliced from top to bottom until loop-C closure induced by GABA in each β3 subunit is shown. Since PTX is bound to the receptor in both structures, the TMDs remain in a closed conformation, limiting the rotation of extracellular domains. This view is followed by a close-up of the β3+/α1− interfaces showing amino acid side chain rearrangements associated with GABA binding. The β3-E+/α1-D− interface is more sensitive to GABA, leading to a higher degree of interface closure and ECD rotation.
Video 6
Bicuculline binding sites in the human α1β3γ2L GABAAR. Bicuculline (BCC) molecules occupy the orthosteric pockets at the β+/α− interfaces (β3-B+/α1-A− is shown). BCC is in ball-and-stick representation (carbon atoms pink; nitrogen, blue; oxygen, red). Side chains of residues proximal to BCC are shown as sticks (nitrogen, blue; oxygen, red) and numbered. The cryo-EM density of BCC is shown as a grey surface.
Video 7
Structure of the human α1β3γ2L GABAAR in complex with alprazolam (Xanax) and GABA. The video first shows a 360° rotation of the GABAAR cryo-EM map (α1, red; β3, blue; γ2, yellow; glycan, orange) in complex with Mb38 (green), phosphatidylinositol 4,5-bisphosphate (PIP2, light brown), alprazolam (ALP, blue) and GABA (khaki) parallel to the plane of the membrane. Then, the cryo-EM density is gradually sliced from top to bottom, revealing the orthosteric binding sites occupied by GABA and the benzodiazepine site occupied by alprazolam. Slicing through the cryo-EM map continues to show the 9' Leu ring and 2' ring residue densities.
Video 8
Alprazolam (Xanax) and diazepam (Valium) binding modes in the α1β3γ2L GABAAR benzodiazepine pocket. ALP and diazepam (DZP) binding to the canonical benzodiazepine pocket at the α1-D+/γ2-C− interface is shown. ALP and DZP are represented as sticks (carbon atoms in blue and teal, respectively; nitrogen, blue; oxygen, red; chlorine, green). Side chains of residues proximal to ALP/DZP are also in stick representation (nitrogen, blue; oxygen, red) and numbered. Cryo-EM densities of ALP/DZP are shown as grey surfaces.
Video 9
Secondary (low affinity) binding site of diazepam (Valium) in the human α1β3γ2L GABAAR transmembrane region. Depiction of diazepam bound to its secondary sites in the putative general anaesthetic pockets at the β+/α− interfaces (β3-E+/α1-D− is shown). Diazepam is in stick representation (carbon atoms teal; nitrogen, blue; oxygen, red; chlorine, green). Side chains of residues proximal to DZP are also shown as sticks and numbered (nitrogen, blue; oxygen, red; sulphur, yellow). The cryo-EM density of DZP is shown as a grey surface.
Video 10
Conformational differences between picrotoxin- (closed/resting) and alprazolam/GABA-bound (desensitized) human α1β3γ2L GABAAR structures. The video shows a morph between PTX- and ALP/GABA-bound α1β3γ2L receptor structures, globally aligned using their TMDs. First, the morph is sliced from top to bottom until loops-C are seen. ALP and GABA binding followed by loop-C closure in both β3 subunits and the concerted ECD rotation is shown. The absence of PTX allows the TMDs to reach a desensitized state. This sequence is followed by side-on views of each receptor interface, alternating between PTX- and ALP/GABA-bound states. This highlights how rotating ECDs communicate to the TMDs through interactions between the β1-β2 and M2-M3 loops.
Rights and permissions
About this article
Cite this article
Masiulis, S., Desai, R., Uchański, T. et al. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature 565, 454–459 (2019). https://doi.org/10.1038/s41586-018-0832-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0832-5
This article is cited by
-
Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment
Nature Communications (2024)
-
A release of local subunit conformational heterogeneity underlies gating in a muscle nicotinic acetylcholine receptor
Nature Communications (2024)
-
Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel
Nature Communications (2024)
-
An Update on Stiripentol Mechanisms of Action: A Narrative Review
Advances in Therapy (2024)
-
Neurosteroids and their potential as a safer class of general anesthetics
Journal of Anesthesia (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.