Abstract
Spliceosomal introns are ubiquitous non-coding RNAs that are typically destined for rapid debranching and degradation. Here we describe 34 excised introns in Saccharomyces cerevisiae that—despite being rapidly degraded in log-phase growth—accumulate as linear RNAs under either saturated-growth conditions or other stresses that cause prolonged inhibition of TORC1, which is a key integrator of growth signalling. Introns that become stabilized remain associated with components of the spliceosome and differ from other spliceosomal introns in having a short distance between their lariat branch point and 3′ splice site, which is necessary and sufficient for their stabilization. Deletion of these unusual introns is disadvantageous in saturated conditions and causes aberrantly high growth rates in yeast that are chronically challenged with the TORC1 inhibitor rapamycin. The reintroduction of native or engineered stable introns suppresses this aberrant rapamycin response. Thus, excised introns function within the TOR growth-signalling network of S. cerevisiae and, more generally, excised spliceosomal introns can have biological functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data and the processed data for each gene and intron are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE121765. For gel source data, see Supplementary Fig. 1. All other data are available from the corresponding author upon reasonable request.
References
Koonin, E. V. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol. Direct 1, 22 (2006).
Irimia, M. & Roy, S. W. Origin of spliceosomal introns and alternative splicing. Cold Spring Harb. Perspect. Biol. 6, a016071 (2014).
Nilsen, T. W. & Graveley, B. R. Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010).
Cech, T. R. & Steitz, J. A. The noncoding RNA revolution–trashing old rules to forge new ones. Cell 157, 77–94 (2014).
Grabowski, P. J., Padgett, R. A. & Sharp, P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell 37, 415–427 (1984).
Padgett, R. A., Konarska, M. M., Grabowski, P. J., Hardy, S. F. & Sharp, P. A. Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors. Science 225, 898–903 (1984).
Ruskin, B., Krainer, A. R., Maniatis, T. & Green, M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38, 317–331 (1984).
Chapman, K. B. & Boeke, J. D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65, 483–492 (1991).
Pai, A. A. et al. The kinetics of pre-mRNA splicing in the Drosophila genome and the influence of gene architecture. eLife 6, e32537 (2017).
Hesselberth, J. R. Lives that introns lead after splicing. Wiley Interdiscip. Rev. RNA 4, 677–691 (2013).
Spingola, M., Grate, L., Haussler, D. & Ares, M., Jr. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5, 221–234 (1999).
Davis, C. A., Grate, L., Spingola, M. & Ares, M., Jr. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28, 1700–1706 (2000).
Juneau, K., Palm, C., Miranda, M. & Davis, R. W. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc. Natl Acad. Sci. USA 104, 1522–1527 (2007).
Ng, R., Domdey, H., Larson, G., Rossi, J. J. & Abelson, J. A test for intron function in the yeast actin gene. Nature 314, 183–184 (1985).
Parenteau, J. et al. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol. Biol. Cell 19, 1932–1941 (2008).
Hooks, K. B., Naseeb, S., Parker, S., Griffiths-Jones, S. & Delneri, D. Novel intronic RNA structures contribute to maintenance of phenotype in Saccharomyces cerevisiae. Genetics 203, 1469–1481 (2016).
Parenteau, J. et al. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147, 320–331 (2011).
Bonnet, A. et al. Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol. Cell 67, 608–621.e606 (2017).
Petfalski, E., Dandekar, T., Henry, Y. & Tollervey, D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18, 1181–1189 (1998).
Gray, J. V. et al. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68, 187–206 (2004).
Wu, T.-T., Su, Y.-H., Block, T. M. & Taylor, J. M. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J. Virol. 70, 5962–5967 (1996).
Lorsch, J. R., Bartel, D. P. & Szostak, J. W. Reverse transcriptase reads through a 2′-5′ linkage and a 2′-thiophosphate in a template. Nucleic Acids Res. 23, 2811–2814 (1995).
Fourmann, J. B. et al. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 27, 413–428 (2013).
Martin, A., Schneider, S. & Schwer, B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277, 17743–17750 (2002).
Katz, Y., Wang, E. T., Airoldi, E. M. & Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 (2010).
LaCava, J. et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 (2005).
Qin, D., Huang, L., Wlodaver, A., Andrade, J. & Staley, J. P. Sequencing of lariat termini in S. cerevisiae reveals 5′ splice sites, branch points, and novel splicing events. RNA 22, 237–253 (2016).
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Jorgensen, P. et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18, 2491–2505 (2004).
Goranov, A. I. et al. Changes in cell morphology are coordinated with cell growth through the TORC1 pathway. Curr. Biol. 23, 1269–1279 (2013).
Zaragoza, D., Ghavidel, A., Heitman, J. & Schultz, M. C. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18, 4463–4470 (1998).
Lempiäinen, H. et al. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell 33, 704–716 (2009).
Mülleder, M. et al. Functional metabolomics describes the yeast biosynthetic regulome. Cell 167, 553–565.e12 (2016).
Rousseau, A. & Bertolotti, A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536, 184–189 (2016).
Aronova, S., Wedaman, K., Anderson, S., Yates, J., III & Powers, T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2779–2794 (2007).
Zurita-Martinez, S. A., Puria, R., Pan, X., Boeke, J. D. & Cardenas, M. E. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics 176, 2139–2150 (2007).
Vyas, V. K., Barrasa, M. I. & Fink, G. R. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 1, e1500248 (2015).
Vyas, V. K. et al. New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi. MSphere 3, e00154-18 (2018).
Munding, E. M., Shiue, L., Katzman, S., Donohue, J. P. & Ares, M. Jr. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell 51, 338–348 (2013).
Warner, J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 (1999).
Gasch, A. P. et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000).
Pleiss, J. A., Whitworth, G. B., Bergkessel, M. & Guthrie, C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell 27, 928–937 (2007).
Bergkessel, M., Whitworth, G. B. & Guthrie, C. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA 17, 1461–1478 (2011).
Zheng, X.-F., Florentino, D., Chen, J., Crabtree, G. R. & Schreiber, S. L. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82, 121–130 (1995).
Weinberg, D. E. et al. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Reports 14, 1787–1799 (2016).
Harju, S., Fedosyuk, H. & Peterson, K. R. Rapid isolation of yeast genomic DNA: Bust ’n Grab. BMC Biotechnol. 4, 8 (2004).
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Wu, X. & Bartel, D. P. kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res. 45, W534–W538 (2017).
Pall, G. S., Codony-Servat, C., Byrne, J., Ritchie, L. & Hamilton, A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 35, e60 (2007).
Chee, M. K. & Haase, S. B. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3 (Bethesda) 2, 515–526 (2012).
Zalatan, J. G. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350 (2015).
McFarlane, E. S. Ribonuclease activity during G1 arrest of the yeast Saccharomyces cerevisiae. Arch. Microbiol. 124, 243–247 (1980).
Wan, R., Yan, C., Bai, R., Lei, J. & Shi, Y. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell 171, 120–132.e112 (2017).
Acknowledgements
We thank A. Amon, C. Burge, D. Pincus, D. Sabatini, P. Sharp and members of the Bartel and Fink laboratories for comments and discussion, G. Li, T. Pham and A. Symbor-Nagrabska for experimental assistance, A. Amon, R. Loewith, S. Schreiber and V. Vyas for reagents, and Whitehead core facilities for sequencing and mass spectrometry. This work was supported by NIH grants GM035010 (G.R.F.) and GM118135 (D.P.B.). D.P.B. is an investigator of the Howard Hughes Medical Institute.
Reviewer information
Nature thanks T. Johnson, S. Montgomery and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Apart from tetrad dissections, which were performed by G.R.F., J.T.M. performed all experiments and analyses. D.P.B. supervised with help from G.R.F. All authors contributed to study design and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 RNA-seq coverage across the branch point and to the 3′SS of the ECM33 intron.
The pileup of reads mapping to the 61-nucleotide region centred on the ECM33 branch point, replotted from Fig. 1b, is shown (red, top). The position of the branch point (closed circle with flanking dashed lines) is indicated on the intron (thick grey line) and relative to the 3′ exon (grey box). Bottom, all reads mapping uniquely to this region are shown (thin grey lines). Reads mapping across the exon–exon junction are coloured blue in the region of the excised intron and are shown above the other reads. Mismatched nucleotides within reads are indicated with coloured bars, with the colour of the bar indicating the identity of the mismatch. Terminal untemplated nucleotides have been clipped from reads.
Extended Data Fig. 2 Stable-intron sedimentation and expression constructs for pull-down and mass spectrometry.
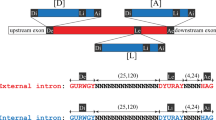
a, Co-sedimentation of the ECM33 stable intron with large ribonucleoprotein complexes. Cleared lysate from a saturated yeast culture was fractionated by sedimentation into a 10–30% sucrose gradient, and RNA was extracted from each fraction. An RNA blot that resolved 25% of the RNA from each fraction and was probed for the ECM33 intron is shown. Fractions are oriented with increasing sedimentation from left to right. RNA was also extracted from 12.5% of the total lysate before sedimentation, of which 50% was loaded for comparison (left lane). The sedimentation of 40S and 60S complexes is marked on the basis of the location of ribosome subunits. Migration of markers with lengths indicated (in nucleotides) is shown on the left. n = 6 biological replicates. b, Behaviour of stable introns ectopically expressed from the URA3 gene. An RNA blot probed for the ECM33 intron after resolving total RNA from cultures expressing the ECM33 intron from the endogenous ECM33 locus (WT, left lanes), cultures from a strain in which the intron had been deleted (ECM33Δintron, middle lanes) and cultures of this deletion strain ectopically expressing the ECM33 intron spliced from the plasmid-borne URA3 reporter gene (right lanes) is shown. pRS416-ECM33_intron plasmid shown on right; PURA3, URA3 promoter; CEN/ARS (centromere/autonomously replicating sequence), low-copy origin of replication. Interior lanes that were not relevant to this experiment were removed from this image (vertical line). Otherwise, this panel is as in Fig. 1c. n = 4 biological replicates. c, Behaviour of stable introns with MS2 hairpins inserted to be used as affinity tags for pull-downs. Five different plasmids with a common backbone (right; PTEF1, TEF1 promoter; FLAG–MCP, coding region of Flag-tagged MS2 coat protein) each expressed URA3 with a different variant of the ECM33 intron (variants A–E, schematized below). These plasmids were each expressed in the strain that lacked an endogenous ECM33 intron (ECM33Δintron). The RNA blot resolved total RNA from the indicated cultures and was probed for a sequence common to the intron variants; otherwise, as in Fig. 1c. The 2 × MS2 hairpin region is 90 nucleotides long, and the expected linear-intron sizes were: A, no intron; B, 330 nucleotides; C, 420 nucleotides; D, 300 nucleotides; and E, 420 nucleotides. n ≥ 2 biological replicates.
Extended Data Fig. 3 Stable introns have oligoadenylate tails.
a, Example of RNA-seq support for intron accumulation and terminal adenylylation. The diagram (top, left) classifies the RNA-seq reads that derive from the possible intron states of ECM33 transcripts (black lines, reads; red dashed lines, intron boundaries). Read counts from log-phase and saturated cultures (normalized for library depth) are listed for each class of reads (top, right). For convenient comparison to intron accumulation, exon reads are only counted if they map within 165 nucleotides of either splice site (thereby encompassing 330 nucleotides, the length of the intron). The alignment (bottom) shows representative reads mapping to the intron 3′ terminus, aligned below the sequence of the 3′ intron–exon junction. Many of these reads had untemplated terminal adenosine residues (blue). b, Composition of untemplated tailing nucleotides observed in a saturated culture. Reads that had at least one terminal untemplated nucleotide after trimming the 3′ adaptor sequence were collected, and the position of this tail was annotated as that of the last templated nucleotide. Counts of reads with tails added at positions 0 to +20 relative to the 5′SS, −10 to +10 relative to the branch point, and −20 to 0 relative to the 3′SS are plotted, binning counts for tails of only adenosines (An, teal) separately from those of all other tails (other, purple). For An tails that map to the 3′-terminal nucleotide of introns, the fraction with each indicated length is plotted (right). The relative abundance of An at positions −2 and −1 relative to the 3′SS was ambiguous because most introns have an adenosine at position −1 (3′SS consensus sequence, YAG), which causes tails of length n at position −1 to be indistinguishable from tails of length n + 1 at position −2.
Extended Data Fig. 4 RNA-seq profiles of genes with stable introns.
Profiles from the stable-intron-inducing condition (red) and log-phase culture (blue) are shown for the indicated 32 genes with confidently identified stable introns not already depicted in Fig. 1. For all but four of these, the profile of the stable-intron-inducing condition is from RNA-seq of the saturated-liquid sample. The four exceptions were not confidently identified in saturated liquid (Extended Data Table 2); for these, the profile of the stable-intron-inducing condition is from either the rapamycin-treated sample (TDA5 and YOS1) or the saturated lawn (RPO26 and RPS8B). Scale bars, 100 nucleotides. Otherwise, this panel is as in Fig. 1a.
Extended Data Fig. 5 Stable introns are indistinguishable from other introns in many respects.
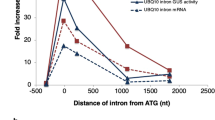
a, Similar splicing motifs compared to other introns. Information-content logos of splicing motifs (6-mer 5′SS, 8-mer branch point and 3-mer 3′SS) for stable introns (bottom) and other introns (top) are plotted. b, Similar length distribution compared to other introns. Cumulative distributions of intron lengths are plotted (P = 0.057, two-tailed Kolmogorov–Smirnov test). c, No significantly enriched motifs within stable introns. Stable-intron sequence motifs discovered by MEME53 are plotted, either controlling for k-mer frequencies of non-stable introns (top) or without controlling (bottom). No significant motifs were discovered in stable introns when k ≥ 6. The motifs discovered without controlling for k-mer frequencies matched the canonical branch point and 5′SS motifs (see a). Branch point and 5′SS motifs were also the only significantly enriched motifs discovered when k ≤ 5. d, No enriched positional k-mer motifs detected by kpLogo54 in stable introns. The most enriched k-mers at positions relative to 5′SS (top), branch point (middle), and 3′SS (bottom) are plotted, comparing stable introns to unstable introns. Stacked nucleotides at a position represent the most significant motif starting at that position. The height is scaled relative to the significance of the motif, as determined by the one-sided binomial test statistic (y axes). Black numbers indicate invariant nucleotides occurring >95% of the time at the position. No k-mers were significantly enriched when using a Bonferroni-corrected P of 0.01. e, Support for a role of branch point–3′SS distance in specifying stable-intron formation. The indicated introns were ectopically expressed from the URA3 splicing construct (Extended Data Fig. 2b). Results are shown from an RNA blot that resolved total RNA from cultures overexpressing the indicated introns and was probed for the indicated introns (length of excised intron in parentheses). The ECM33 and RPL27B introns were probed sequentially, and then the ACT1, HNT2 and APS3 introns were probed concurrently. ACT1 probe was validated on synthetic transcripts resembling the ACT1 intron, which were produced by in vitro transcription (not shown). Migration of markers with lengths (in nucleotides) indicated is at the left. n ≥ 2 biological replicates. f, Expression of mRNAs from genes containing stable introns. Left, relationship between the expression in log-phase and saturated cultures (expression cutoff, 1 TPM in both samples). Points for genes containing stable introns are indicated (orange), and the point for ECM33 is labelled. Right, comparison of the expression results of genes with stable introns to those of the remaining genes. The cumulative distributions of fold changes in expression (expressed as log2(mRNA in log-phase culture/mRNA in saturated culture)) observed between log-phase and saturated cultures are plotted, which shows no significant difference between stable-intron genes and other genes (P = 0.31 two-tailed Kolmogorov–Smirnov test).
Extended Data Fig. 6 Assessing aspects of TORC1 regulation on stable-intron expression.
a, Inability of TORC1-independent inhibition of protein synthesis to prematurely induce stable-intron accumulation. The left lanes show a replicate of Fig. 3b, and the right lanes show results after treatment with either low (0.25 mg l−1) or high (25 mg l−1) concentrations of cycloheximide. As indicated by OD600 nm at collection, the mild cycloheximide treatment allowed the culture to reach an OD600 nm of 3.4 after 24 h, which was equivalent to the OD600 nm of 10 h without cycloheximide (Fig. 3a). b, Dispensability of TORC1-responsive Tap42 for stable-intron formation. Samples were grown at 25 °C owing to temperature sensitivity of the tap42-11 allele. Additionally, owing to slower growth at 25 °C, the duration of pre-growth was extended to 5.5 h before reseeding in medium with (+) or without (−) rapamycin, such that the change in OD600 nm of the pre-growth sample matched that of wild-type cultures grown at 30 °C for 4 h. Otherwise, this panel is as in Fig. 3b. n = 2 biological replicates. c, Dispensability of TORC1-sensitive Sch9 for stable-intron formation. Samples were grown in SC–Trp medium to maintain plasmids that expressed either TORC1-sensitive (pSCH9) or TORC1-insensitive (pSCH9DE) Sch9 protein. Because of the slower growth of the sch9Δ pRS414::SCH9 and sch9Δ pRS414::SCH9DE strains (possibly owing to the requisite SC–Trp medium) the duration of pre-growth was extended to 5 h, such that the change in OD600 nm of the pre-growth sample matched that of wild-type cultures grown for 4 h. Otherwise, this panel is as in Fig. 3b. n = 3 biological replicates. d, Dispensability of Sch9 for stable-intron formation. The left lanes show a replicate of Fig. 3b, and the right lanes show the same experiments performed in the sch9Δ strain. n = 2 biological replicates (sch9Δ samples). e, Inability of acute deprivation of select nutrients to induce accumulation of stable introns. To prevent contamination of starting cultures with stable introns contributed by inoculum, cultures were seeded at OD600 nm = 0.2 from an overnight culture that was allowed to grow to mid-log phase, collected by vacuum filtration, washed in water and resuspended in fresh medium that lacked the indicated nutrients. Cultures were collected after the indicated times and analysed as in Fig. 1c, except the RNA blot was sequentially re-probed for the ECM33, SAC6 and USV1 introns. As indicated by OD600 nm at collection, the sample deprived of ammonium sulfate (the main source of nitrogen) was still able to reach a high density after 24 h. n ≥ 2 biological replicates. f, Inability of the general amino acid control response to induce stable introns. Early-log-phase cultures were treated with either 100 nM rapamycin or 50 mM 3-aminotriazole for the indicated times. Cultures were grown in SC–His to force inhibition of histidine biosynthesis by 3-aminotriazole. Cultures treated with 3-aminotriazole for 24 h did not reach a density sufficient to induce stable-intron accumulation by approaching saturated growth. g, Stable-intron accumulation despite inability to induce the general amino acid control pathway. The left lanes show a replicate of Fig. 3b, and the middle and right lanes show the same experiments performed in gcn2Δ and gcn4Δ strains, respectively.
Extended Data Fig. 7 The influence of hyperactive TORC1 on stable-intron formation.
a, The left and right panels are biological replicates comparing ECM33 and SAC6 intron levels in a strain with hyperactive TORC1 (TORC1*) to those in a strain-background control (cdc28-4). Growth conditions were as described for these strains31. Overexpression of Sfp1 was induced 1 h before treatment with rapamycin (or no treatment). Owing to the decreased growth rate of these strains in the requisite conditions, samples with and without rapamycin were collected after 6 h of treatment rather than after 4 h of treatment. Numbers below the ECM33 and SAC6 intron blots indicate the level of each intron normalized to the level in the cdc28-4 rapamycin-treated samples, after first normalizing all lanes to the 5.8S rRNA loading control (set to 1). b, Accumulation of the ECM33 intron in rapamycin-treated, saturated-lawn and saturated-liquid cultures, detected using RNA-seq. Results showing accumulation of this intron in saturated-liquid culture but not log-phase culture are from a different biological replicate than that shown in Fig. 1a, b; otherwise, this panel is as in Fig. 1a. Rapamycin results are re-plotted from Fig. 3d (4 h). c, Overlap between stable introns identified in saturated-liquid, saturated-lawn and rapamycin-treated cultures.
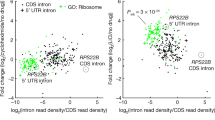
Extended Data Fig. 8 Evidence for spliceosome sequestration and control of ribosome production by stable introns.
a, No evidence for compensatory stable-intron expression after genomic deletion of five stable introns. The scatter plot shows the relationship between intron accumulation in rapamycin-treated, wild-type culture and in rapamycin-treated, EUHSR culture, focusing on the 25 stable introns detected after rapamycin treatment (Extended Data Table 2). The dashed line is placed at x = y. Stable introns are indicated (closed orange circles). Points for introns deleted from the EUHSR genome (ECM33, UBC4, HNT1, SAC6 and RFA2) are labelled. All introns were pseudocounted at 0.5 TPM. b, Aggregate stable-intron accumulation approaching that of spliceosomal RNAs. Levels of stable introns and spliceosomal RNAs (labelled closed blue circles) are plotted, comparing levels in log-phase, wild-type culture to those in rapamycin-treated, wild-type culture. The aggregate stable-intron abundance (closed orange circle, stable intron total) is also plotted. Much of the apparent accumulation observed in log-phase culture was attributable to reads from a few highly transcribed stable-intron genes (for example, RPS29A, which has an intron:mRNA TPM ratio of 107:2,030 in log phase compared to 289:319 in saturated culture). Otherwise, as in a. c, Reduced expression of mRNA of ribosomal protein genes (RPGs) when overexpressing a stable intron in rapamycin-treated EUHSR culture. The cumulative distributions of fold changes in mRNA expression (expressed as log2(mRNA in EUHSR expressing ECM33 intron/mRNA in EUHSR expressing ACT1 intron)) observed between an EUHSR culture with stable-intron (ECM33) ectopic expression and an EUHSR culture with control-intron (ACT1) ectopic expression are plotted. The distribution of changes for mRNAs of RPGs (green) differed from that of other genes (black), with generally lower expression of RPGs in the culture that overexpresses the stable intron (P = 8.8 × 10−32, one-tailed Kolmogorov–Smirnov test; expression cutoff, 1 TPM in both samples). d, Less efficient splicing—as detected by increased intron retention—when overexpressing a stable intron in rapamycin-treated, EUHSR culture. When analysing the dataset in c, 103 genes had significantly more intron retention when ectopically expressing the stable intron compared to when expressing the control intron (MISO, Bayes factor > 100). The Venn diagram shows the overlap between these genes with increased intron retention and intron-containing RPGs (P = 3.9 × 10−22, hypergeometric test).
Supplementary information
Supplementary Figure 1
This file contains the uncropped RNA blots.
Supplementary Table 1
A list of S. cerevisiae strains used and generated in this study.
Supplementary Table 2
A list of plasmids used and generated in this study.
Supplementary Table 3
A list of CRISPR guide and repair oligos.
Supplementary Table 4
A list of RNA-blot probes.
Supplementary Table 5
A list of mutant intron sequences.
Rights and permissions
About this article
Cite this article
Morgan, J.T., Fink, G.R. & Bartel, D.P. Excised linear introns regulate growth in yeast. Nature 565, 606–611 (2019). https://doi.org/10.1038/s41586-018-0828-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0828-1
This article is cited by
-
NAP-seq reveals multiple classes of structured noncoding RNAs with regulatory functions
Nature Communications (2024)
-
Nutrient-dependent regulation of a stable intron modulates germline mitochondrial quality control
Nature Communications (2024)
-
Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects
Nature Reviews Genetics (2023)
-
Single-cell analysis reveals region-heterogeneous responses in rhesus monkey spinal cord with complete injury
Nature Communications (2023)
-
Novel single nucleotide polymorphisms of insulin-like growth factor-binding protein 7 (IGFBP7) gene significantly associated with growth traits in striped catfish (Pangasianodon hypophthalmus Sauvage, 1878)
Molecular Genetics and Genomics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.