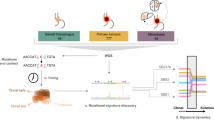
Abstract
Clonal expansion in aged normal tissues has been implicated in the development of cancer. However, the chronology and risk dependence of the expansion are poorly understood. Here we intensively sequence 682 micro-scale oesophageal samples and show, in physiologically normal oesophageal epithelia, the progressive age-related expansion of clones that carry mutations in driver genes (predominantly NOTCH1), which is substantially accelerated by alcohol consumption and by smoking. Driver-mutated clones emerge multifocally from early childhood and increase their number and size with ageing, and ultimately replace almost the entire oesophageal epithelium in the extremely elderly. Compared with mutations in oesophageal cancer, there is a marked overrepresentation of NOTCH1 and PPM1D mutations in physiologically normal oesophageal epithelia; these mutations can be acquired before late adolescence (as early as early infancy) and significantly increase in number with heavy smoking and drinking. The remodelling of the oesophageal epithelium by driver-mutated clones is an inevitable consequence of normal ageing, which—depending on lifestyle risks—may affect cancer development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All the WES, WGS and SNP array data have been deposited in the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/) under accession numbers EGAS00001003008, EGAS00001003281 and EGAS00001003331, respectively. Data for Figures and Extended Data Figures are available as Source Data. All other data are available from the corresponding author on reasonable request.
References
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Cooper, C. S. et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 47, 367–372 (2015).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041 (2017).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Pickens, A. & Orringer, M. B. Geographical distribution and racial disparity in esophageal cancer. Ann. Thorac. Surg. 76, S1367–S1369 (2003).
Enzinger, P. C. & Mayer, R. J. Esophageal cancer. N. Engl. J. Med. 349, 2241–2252 (2003).
Matejcic, M., Gunter, M. J. & Ferrari, P. Alcohol metabolism and oesophageal cancer: a systematic review of the evidence. Carcinogenesis 38, 859–872 (2017).
Ishiguro, S. et al. Effect of alcohol consumption, cigarette smoking and flushing response on esophageal cancer risk: a population-based cohort study (JPHC study). Cancer Lett. 275, 240–246 (2009).
Katada, C. et al. Alcohol consumption and multiple dysplastic lesions increase risk of squamous cell carcinoma in the esophagus, head, and neck. Gastroenterology 151, 860–869.e7 (2016).
Tian, D. et al. Multifocal accumulation of p53 protein in esophageal carcinoma: evidence for field cancerization. Int. J. Cancer 78, 568–575 (1998).
Chai, H. & Brown, R. E. Field effect in cancer–an update. Ann. Clin. Lab. Sci. 39, 331–337 (2009).
Sawada, G. et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology 150, 1171–1182 (2016).
Cancer Genome Atlas Research Network. Integrated genomic characterization of oesophageal carcinoma. Nature 541, 169–175 (2017).
Chang, J. et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat. Commun. 8, 15290 (2017).
Gao, Y. B. et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102 (2014).
Qin, H. D. et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am. J. Hum. Genet. 98, 709–727 (2016).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Totoki, Y. et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 46, 1267–1273 (2014).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Bulavin, D. V. et al. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16Ink4a–p19Arf pathway. Nat. Genet. 36, 343–350 (2004).
Hori, M. et al. Cancer incidence and incidence rates in Japan in 2009: a study of 32 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn. J. Clin. Oncol. 45, 884–891 (2015).
Demehri, S., Turkoz, A. & Kopan, R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell 16, 55–66 (2009).
Li, Z. S. & Li, Q. The latest 2010 WHO classification of tumors of digestive system. Zhonghua Bing Li Xue Za Zhi 40, 351–354 (2011).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Kataoka, K. et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 534, 402–406 (2016).
Shiraishi, Y. et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 41, e89 (2013).
Suzuki, H. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Nannya, Y. et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 65, 6071–6079 (2005).
Yamamoto, G. et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am. J. Hum. Genet. 81, 114–126 (2007).
Yoshizato, T. et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood 129, 2347–2358 (2017).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Shiraishi, Y., Tremmel, G., Miyano, S. & Stephens, M. A simple model-based approach to inferring and visualizing cancer mutation signatures. PLoS Genet. 11, e1005657 (2015).
Blokzijl, F., Janssen, R., Van Boxtel, R. & Cuppen, E. MutationalPatterns: an integrative R package for studying patterns in base substitution catalogues. Preprint at https://www.biorxiv.org/content/early/2016/08/30/071761 (2016).
Roth, A. et al. PyClone: statistical inference of clonal population structure in cancer. Nat. Methods 11, 396–398 (2014).
Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48 (2010).
Acknowledgements
We thank the Ministry of Education, Culture, Sports, Science and Technology for support (grant references: hp150232 and 15H05909), together with many other funding bodies and individuals (Supplementary Note 1).
Reviewer information
Nature thanks F. Ciccarelli, B. Lehner and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.Y., H. Suzuki, T.Y., N.K., H.M., M.M. and S. Ogawa designed the study. S. Ohashi, Y.A., I.A., T. Horimatsu, S. Miyamoto, S.T., Y. Sakai, Yoshitaka Sato, H. Seno, M.M. and Y. Nishikawa provided specimens. Y.T., S. Minamiguchi and H.H. performed histological analysis. A.Y., H. Suzuki, T.Y., N.K., Y. Shiozawa, Yusuke Sato, K.A., Y.T., S.K.K., Y.F., K.Y., K.K., Y.I., T. Hirano and M.M.N. performed sample preparation. G.S. and K.M. provided sequencing data. M.S., A.Y., H. Suzuki, T.Y., N.K., Y. Shiozawa and Y. Nannya performed mutation calling, validated the results, and analysed copy-number alterations, mutational signature and clonal dynamics. A.Y., H. Suzuki, T.Y., N.K., Y. Shiozawa, K.A., Y.T., Y. Nannya, Y. Shiraishi, K.C., H.T., M.N., J.B.B. and S. Miyano performed bioinformatics analysis. A.Y., H. Suzuki, T.Y., N.K., Y. Nannya, H.M. and S. Ogawa prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Study design.
a–c, Summary of the subjects, samples, methods of sample collection, lifestyle ESCC risks, and histology, as well as sequencing platforms used for analysis of solitary samples (a), single-cell-derived colonies (b) and densely collected samples (c). *In one case (UPN53), only cancer and dysplasia samples were collected. §, UPN36, UPN37 and UPN39; ¶, UPN49; ‡, UPN88; #, UPN56 and UPN61; Φ, UPN58; ††, UPN55, UPN59, UPN60 and UPN65. Samples from these individuals were analysed in indicated multiple experiments (see a, b and c). †Out of 53 PNE samples, 11 were analysed in both solitary and high-density sampling (see a and c).
Extended Data Fig. 2 WES of PNE and cancer samples from a surgical specimen.
a, Positions of 25 PNE and 2 cancer samples from a surgically resected oesophagus of a 58-year-old, high-risk patient with ESCC (UPN32) are shown by green and red circles, respectively. Calibration is shown on the right. EGJ, oesophagogastric junction. b, MCFs are compared between samples from PNE (green) (n = 25) and cancer (red) (n = 2) samples. c, d, MCF is plotted for all shared (orange) and private (grey) mutations for low-density (c) and high-density (d) sampling. Driver mutations are indicated by arrowheads in indicated colours. The position of each sample is shown on the left. Calibration for the number of mutations depicted in the horizontal axis is separately provided for each sampling. e, For preparation of PNE samples, after submucosal layers were removed from endoscopic biopsy specimens, 0.2-mm2, 0.8-mm2 or 4.0-mm2 samples were collected using a punch biopsy of 0.5 mm or 1.0 mm in diameter, or 2 × 2-mm rectangular excision, respectively, followed by DNA extraction. f, Photographs of punch biopsy devices for 0.5-mm-diameter and 1.0-mm-diameter sampling. Photographs copyright Kai Industries. g, Histogram of DNA recovered from 0.2-mm2, 0.8-mm2 or 4.0-mm2 samples. Box plots represent the median, first quartile and third quartile, with whiskers extending to the furthest value within 1.5× interquartile range.
Extended Data Fig. 3 Somatic mutations in solitary sampling.
a, Summary of somatic mutations found in 157 PNE, 12 dysplasia and 20 cancer samples from individuals with (pink) or without (green) lifestyle ESCC risks. MCFs are plotted for all mutations (upper panel). Driver mutations are shown by coloured triangles as indicated. Information about histology and sample size is shown on the top lanes. The age of the subject and the total number of mutations are shown in the bottom panels. Status of ESCC, CNAs and mutations are also shown (middle panels). b–j, MCFs of somatic mutations detected in WES of multiple samples from PNE, as well as dysplastic and cancer tissues, are depicted separately for shared (orange) and non-shared mutations (grey) in representative cases from 16 independent individuals, for whom multi-regional sampling was performed. Mutations in TP53, NOTCH1, PPM1D and other drivers are indicated by arrowheads in different colours. Positions of tumours and biopsy samples are shown on the left in each panel. Distance from the maxillary dental arch is indicated. Mutations are shared only between cancer samples except in two cases (i and j), in which small numbers of mutations (between 3 and 16) are shared between mutually positioned samples as indicated.
Extended Data Fig. 4 Signatures of mutations in PNE, dysplasia and cancer samples.
a, Number (middle) and relative frequency (bottom) of mutations allocated to signatures A–D in the individual samples. Information about pathology, lifestyle ESCC risks, cohort and age of the subject is indicated (top). b, Number of mutations allocated to indicated mutational signatures is plotted against the age of the subject for PNE samples from low-risk (blue) and high-risk (red) individuals. Numbers of samples from low-risk and high-risk individuals are 34 and 19 (0.2-mm2 sampling), 19 and 12 (0.8-mm2 sampling), and 40 and 33 (4-mm2 sampling), respectively. Regression lines for samples from low-risk individuals are indicated with R2. P values for significance between samples from high-risk and low-risk individuals are also provided (one-sided Mann–Whitney U-test). c, Mean (± 95% confidence interval) of standardized residuals of the number of mutations allocated to each signature in samples from high-risk individuals against the linear regression model in samples from low-risk individuals is plotted for 0.2-mm2, 0.8-mm2 and 4-mm2 samples and for combined samples in a random-effects model. P value in the random-effects model is indicated, together with the weight from each sample size during the model fitting (two-sided Wald test) (Methods). d, Mean (±s.d.) number of mutations allocated to signature 16 in oesophagus (ESCC) (n = 90) (left), hepatic (LIHC) (n = 361) (middle) and stomach (STAD) (n = 239) (right) cancers from TCGA, are plotted according to the status of the ALDH2 risk allele. P values for significance between samples from risk-positive and risk-negative individuals are also provided (two-sided Mann–Whitney U-test). e, Frequency of mutations allocated to signature C in Japanese ESCC samples is plotted according to the status of alcohol drinking and smoking. According the history of heavy drinking and smoking, the subjects were divided into four groups; drinking- and smoking-negative (n = 18); drinking-negative and smoking-positive (n = 3); drinking-positive and smoking-negative (n = 40); and drinking- and smoking-positive (n = 96), as indicated in the panel. Box plots represent the median, first quartile and third quartile, with whiskers extending to the furthest value within 1.5× interquartile range; points show outliers.
Extended Data Fig. 5 Mutations detected in single-cell-derived colonies in WGS.
VAF histograms of somatic mutations detected in 13 single-cell-derived colonies analysed with WGS are depicted for each sample. Mutation number and driver genes mutated in each sample are indicated.
Extended Data Fig. 6 Driver mutations in PNE samples.
a, Distribution of mutations in 157 PNE and 519 ESCC samples is shown for driver genes significantly mutated in PNE or ESCC samples, including NOTCH2, NOTCH3, CREBBP, FAT1, CHEK2, PAX9, EP300 and PIK3CA. Mutations are depicted separately for PNE (top) and ESCC (bottom). b, Frequencies of driver genes in paired cancer and PNE samples from 68 patients with ESCC. Genes in which mutations were observed in 5% or more samples either in PNE or ESCC were evaluated. Significantly differentially mutated genes (q < 0.05) between PNE and ESCC samples are indicated by asterisks. c, d, Number of driver mutations (c) and their maximum MCFs (d) in samples from high-risk and low-risk individuals are plotted against the age of subject, according to sample size. Regression lines are provided with R2. P values for the significance of lifestyle ESCC risks are also indicated (one-sided Mann–Whitney U-test). e, h, Mean (± 95% confidence interval) of standardized difference of mutation number (e) and maximum MCF (h) between samples from high-risk and low-risk individuals are plotted for each sampling size, and combined samples, using a random-effects model. P value in the random-effects model is also indicated, together with the weight from each sample size (in per cent) for the model fitting (two-sided Wald test) (Methods). f, g, Mean (± 95% confidence interval) of standardized residuals of the number of indicated driver mutations (f) and their maximum MCFs (g) in samples from high-risk individuals against the linear regression model in samples from low-risk individuals (Methods); plotted for 0.2-mm2, 0.8-mm2 and 4-mm2 samples. Standardized residuals for combined samples using a random-effects model are also shown. P-value in the random-effects model is also indicated, together with the weight from each sample size (in per cent) for the model fitting (two-sided Wald test) (Methods). In the analyses in c–h, the numbers of samples from low-risk and high-risk individuals, respectively, are 34 and 19 (0.2-mm2 sampling), 19 and 12 (0.8-mm2 sampling), and 40 and 33 (4-mm2 sampling) (c, e, f); 26 and 18 (0.2-mm2 sampling), 13 and 12 (0.8-mm2 sampling), and 30 and 31 (4-mm2 sampling) (d, h); 5 and 5 (0.2-mm2 sampling), 4 and 7 (0.8-mm2 sampling), and 8 and 19 (4-mm2 sampling) (TP53 in g); 14 and 18 (0.2-mm2 sampling), 7 and 11 (0.8-mm2 sampling), 21 and 28 (4-mm2 sampling) (NOTCH1 in g); 2 and 4 (0.2-mm2 sampling), 2 and 9 (4-mm2 sampling) (PPM1D in g).
Extended Data Fig. 7 CNAs in PNE, dysplasia and cancer samples.
a, Colour-gradient maps of CNAs and UPDs as detected by SNP-array karyotyping or sequencing-based assays are shown for PNE samples from high-risk and low-risk individuals, as well as dysplastic tissues (n = 12) and cancer samples (n = 51) (middle panels). Fractions of genomes showing copy-number gains (red), losses (blue) and UPD (green) are also plotted (top panels). Information about histology, age of the subject, sample size and risks of developing ESCC are shown in top panels. b, Box plots of fractions of genomic regions showing CNAs in PNE (n = 188), dysplasia (n = 12) and cancer (n = 45) samples; the median, first and third quartiles, as well as outliers, are indicated with whiskers extending to the furthest value within 1.5 of the interquartile range. P values for significant differences are from two-sided Mann–Whitney U-test. DP, dysplasia. c, Effects of lifestyle ESCC risks and age on CNAs. Mean (± 95% confidence interval) of standardized residuals of the total fraction of genomic regions showing CNAs in samples from high-risk individuals, compared with the linear regression model in samples from low-risk individuals (left panel) and Fisher’s z-transformed correlation of age to the total fraction of genomic regions showing CNAs in samples from low-risk individuals (right panel) are plotted for 0.2-mm2, 0.8-mm2, 4-mm2 and 8-mm2 samples, which are combined using a random-effects model. P value in the random-effects model is also indicated, together with the weight from each sample size (in per cent) for the model fitting (two-sided Wald test) (Methods). Numbers of samples from low-risk and high-risk individuals, respectively, are 34 and 19 (0.2-mm2 sampling), 19 and 12 (0.8-mm2 sampling), 40 and 33 (4-mm2 sampling), and 3 and 28 (8-mm2 sampling). Numbers of samples from low-risk individuals who are <50 years old and ≥50 years old, respectively, are 20 and 14 (0.2-mm2 sampling), 13 and 6 (0.8-mm2 sampling), and 7 and 33 (4-mm2 sampling), respectively. d, f, LOH maps of chromosomes 9 (d) and 17 (f) in PNE (top) and cancer (bottom) samples. Deletions and UPDs are shown by blue and green lines, respectively. Positions and mutation status of CDKN2A, NOTCH1 and TP53 genes are indicated. e, g, Bar plots of frequencies of 9q UPD (e) and 17p LOH (g) in samples from high-risk and low-risk individuals. Individuals of <50 years old (low risk, n = 40; high risk, n = 11) and those of ≥50 years old (low-risk, n = 56; high risk, n = 81) were analysed. Number (n) of samples in each group is also indicated. P values are for significant differences between both risk groups (two-sided Fisher’s exact test). Whiskers indicate the upper bounds of 95% confidence intervals from the binomial distribution.
Extended Data Fig. 8 Spatial architecture of clones in representative PNE biopsies.
a, Mutation analysis of 341 samples obtained by high-density sampling from 16 biopsy specimens from 14 individuals with different age, lifestyle ESCC risks, and ESCC status (bottom panels), using WES (249 from 10 biopsies) or targeted-capture sequencing of major drivers (92 from 6 biopsies). Total numbers of mutations (top) are depicted for samples analysed by WES. Shared mutations are shown in orange. Mutation status of major drivers and common CNAs are summarized (middle panels). b–e, MCFs of detected mutations are depicted for each sample obtained by high-density collection from PNE biopsies performed on low-risk healthy individuals in their 20s (b–d) and an 81-year-old, low-risk man (e). Four representative examples from 10 biopsies analysed by high-density WES sampling are shown. Mutations shared by a distinct set of samples or isolated samples are integrated to identify unique clones, as shown in different colours. Driver mutations are indicated by downward-pointing arrowheads. Rightward-pointing arrows indicate the clones in the same colour in Fig. 4b, c and Extended Data Fig. 9a, b.
Extended Data Fig. 9 Clones detected in high-density sampling.
a–i, The spatial distribution of clones as revealed by WES or targeted sequencing of driver genes is depicted for biopsies from younger-aged low-risk (a, b), middle-aged low-risk and high-risk (c, d), 70-year-old low-risk (e, f) and elderly high-risk (g–i) subjects, as described in Fig. 4. For convenience, samples that share discrete sets of mutations—as indicated by different colours—are summarized for four subjects (a–d); in these subjects, mutations are frequently shared between distant samples, which suggests mosaicism. Sequencing platforms are indicated in each panel.
Extended Data Fig. 10 Phylogenetic analysis of representative clones.
Phylogenetic trees mapped to the location of samples are depicted for additional representative clones from a total of 29 clones, seen in 5 biopsies from 5 elderly subjects. Imputed clonal structures are depicted in violin plots at the bottom, in which the estimated cellular prevalence of imputed mutational clusters (vertical axis) in each sample are shown as the distribution of posterior probabilities (width of the violin plots) calculated from the PyClone model. Colours correspond to those in the associated phylogenetic trees.
Supplementary information
Supplementary Information
This file contains Supplementary Methods 1-2 and Supplementary Note 1, which contains the acknowledgements.
Supplementary Tables
This file contains Supplementary Tables 1-10.
Source data
Rights and permissions
About this article
Cite this article
Yokoyama, A., Kakiuchi, N., Yoshizato, T. et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317 (2019). https://doi.org/10.1038/s41586-018-0811-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0811-x
This article is cited by
-
Cell competition and cancer from Drosophila to mammals
Oncogenesis (2024)
-
Regulatory T cell-mediated immunosuppression orchestrated by cancer: towards an immuno-genomic paradigm for precision medicine
Nature Reviews Clinical Oncology (2024)
-
Aneuploidy and complex genomic rearrangements in cancer evolution
Nature Cancer (2024)
-
Decoding the basis of histological variation in human cancer
Nature Reviews Cancer (2024)
-
Somatic mutations in aging and disease
GeroScience (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.