Abstract
Patients with glioblastoma currently do not sufficiently benefit from recent breakthroughs in cancer treatment that use checkpoint inhibitors1,2. For treatments using checkpoint inhibitors to be successful, a high mutational load and responses to neoepitopes are thought to be essential3. There is limited intratumoural infiltration of immune cells4 in glioblastoma and these tumours contain only 30–50 non-synonymous mutations5. Exploitation of the full repertoire of tumour antigens—that is, both unmutated antigens and neoepitopes—may offer more effective immunotherapies, especially for tumours with a low mutational load. Here, in the phase I trial GAPVAC-101 of the Glioma Actively Personalized Vaccine Consortium (GAPVAC), we integrated highly individualized vaccinations with both types of tumour antigens into standard care to optimally exploit the limited target space for patients with newly diagnosed glioblastoma. Fifteen patients with glioblastomas positive for human leukocyte antigen (HLA)-A*02:01 or HLA-A*24:02 were treated with a vaccine (APVAC1) derived from a premanufactured library of unmutated antigens followed by treatment with APVAC2, which preferentially targeted neoepitopes. Personalization was based on mutations and analyses of the transcriptomes and immunopeptidomes of the individual tumours. The GAPVAC approach was feasible and vaccines that had poly-ICLC (polyriboinosinic-polyribocytidylic acid-poly-l-lysine carboxymethylcellulose) and granulocyte–macrophage colony-stimulating factor as adjuvants displayed favourable safety and strong immunogenicity. Unmutated APVAC1 antigens elicited sustained responses of central memory CD8+ T cells. APVAC2 induced predominantly CD4+ T cell responses of T helper 1 type against predicted neoepitopes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw data that support the findings of this study shown in Figs. 2–4 and Extended Data Figs. 1, 3–8 and 10 are available from the corresponding author upon reasonable request. Data in Fig. 5, Extended Data Fig. 9, Table 1 and Supplementary Table 5 are on patient outcomes from the clinical study. These data will be available upon reasonable request to the corresponding author from the GAPVAC-101 study case record forms. Supplementary Tables 1–4 represent raw data and should be used to recapitulate the findings demonstrated in this study. Mass spectrometry data for sequence identification of APVAC warehouse peptides have been deposited at the PeptideAtlas (http://www.peptideatlas.org/) with the dataset identifier PASS01284. mRNA microarray data used for APVAC selection are available at Gene Expression Omnibus (GSE122498). All mutations found in the DNA sequencing are depicted in the manuscript. The uploading of DNA sequencing data with the potential for patient identification was not included in the informed consent that patients gave to the trial; these data will be shared (with identifying information removed) upon reasonable request.
Change history
07 February 2019
The additional author support information was erroneously omitted from the Supplementary Information. This has been corrected online.
References
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010).
Robert, C. et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330 (2015).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Quail, D. F. & Joyce, J. A. The microenvironmental landscape of brain tumors. Cancer Cell 31, 326–341 (2017).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Yadav, M. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014).
Freudenmann, L. K., Marcu, A. & Stevanović, S. Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry. Immunology 154, 331–345 (2018).
Bouffet, E. et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 34, 2206–2211 (2016).
Johanns, T. M. et al. Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 6, 1230–1236 (2016).
Platten, M., Bunse, L., Wick, W. & Bunse, T. Concepts in glioma immunotherapy. Cancer Immunol. Immunother. 65, 1269–1275 (2016).
Okada, H. et al. Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with α-type 1 polarized dendritic cells and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J. Clin. Oncol. 29, 330–336 (2011).
Weller, M. et al. Vaccine-based immunotherapeutic approaches to gliomas and beyond. Nat. Rev. Neurol. 13, 363–374 (2017).
Rampling, R. et al. A Cancer Research UK first time in human phase I trial of IMA950 (novel multi peptide therapeutic vaccine) in patients with newly diagnosed glioblastoma. Clin. Cancer Res. 22, 4776–4785 (2016).
Weinschenk, T. et al. Integrated functional genomics approach for the design of patient-individual antitumor vaccines. Cancer Res. 62, 5818–5827 (2002).
Dutoit, V. et al. Exploiting the glioblastoma peptidome to discover novel tumour-associated antigens for immunotherapy. Brain 135, 1042–1054 (2012).
Morse, M. A. et al. Phase I study utilizing a novel antigen-presenting cell-targeted vaccine with Toll-like receptor stimulation to induce immunity to self-antigens in cancer patients. Clin. Cancer Res. 17, 4844–4853 (2011).
Lawson, D. H. et al. Randomized, placebo-controlled, phase III trial of yeast-derived granulocyte–macrophage colony-stimulating factor (GM-CSF) versus peptide vaccination versus GM-CSF plus peptide vaccination versus placebo in patients with no evidence of disease after complete surgical resection of locally advanced and/or stage IV melanoma: a trial of the Eastern Cooperative Oncology Group-American College of Radiology Imaging Network Cancer Research Group (E4697). J. Clin. Oncol. 33, 4066–4067 (2015).
Woroniecka, K. et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin. Cancer Res. 24, 4175–4186 (2018).
Hadrup, S. R. et al. Tumor infiltrating lymphocytes in seminoma lesions comprise clonally expanded cytotoxic T cells. Int. J. Cancer 119, 831–838 (2006).
Thor Straten, P. et al. Identification of identical TCRs in primary melanoma lesions and tumor free corresponding sentinel lymph nodes. Cancer Immunol. Immunother. 55, 495–502 (2006).
Kalaora, S. et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget 7, 5110–5117 (2016).
Tran, E. et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015).
Weller, M. et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 18, 1373–1385 (2017).
Shraibman, B. et al. Identification of tumor antigens among the HLA peptidomes of glioblastoma tumors and plasma. Mol. Cell. Proteomics 17, 2132–2145 (2018).
Singh-Jasuja, H., Emmerich, N. P. & Rammensee, H. G. The Tübingen approach: identification, selection, and validation of tumor-associated HLA peptides for cancer therapy. Cancer Immunol. Immunother. 53, 187–195 (2004).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Walter, S. et al. Cutting edge: predetermined avidity of human CD8 T cells expanded on calibrated MHC/anti-CD28-coated microspheres. J. Immunol. 171, 4974–4978 (2003).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 (2008).
Kim, Y. et al. Immune epitope database analysis resource. Nucleic Acids Res. 40, W525–W530 (2012).
Kreiter, S. et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015).
Koressaar, T. & Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291 (2007).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Stupp, R., Brada, M., van den Bent, M. J., Tonn, J. C. & Pentheroudakis, G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 25, iii93–iii101 (2014).
Wen, P. Y. et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J. Clin. Oncol. 28, 1963–1972 (2010).
Rodenko, B. et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1, 1120–1132 (2006).
Hadrup, S. R. et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods 6, 520–526 (2009).
Walter, S. et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 18, 1254–1261 (2012).
Bertoletti, A. et al. Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein. J. Virol. 67, 2376–2380 (1993).
Livingston, B. D. et al. The hepatitis B virus-specific CTL responses induced in humans by lipopeptide vaccination are comparable to those elicited by acute viral infection. J. Immunol. 159, 1383–1392 (1997).
Welters, M. J. et al. Harmonization of the intracellular cytokine staining assay. Cancer Immunol. Immunother. 61, 967–978 (2012).
Zhu, J., Yamane, H. & Paul, W. E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010).
Loffler, M. W. et al. Personalized peptide vaccine induced immune response associated with long-term survival of a metastatic cholangiocarcinoma patient. J. Hepatol. 65, 849–855 (2016).
Widenmeyer, M. et al. Promiscuous survivin peptide induces robust CD4+ T-cell responses in the majority of vaccinated cancer patients. Int. J. Cancer 131, 140–149 (2012).
Grimm, E. A. et al. Characterization of interleukin-2-initiated versus OKT3-initiated human tumor-infiltrating lymphocytes from glioblastoma multiforme: growth characteristics, cytolytic activity, and cell phenotype. Cancer Immunol. Immunother. 32, 391–399 (1991).
Britten, C. M. et al. The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol. Immunother. 57, 289–302 (2008).
van der Burg, S. H. et al. Harmonization of immune biomarker assays for clinical studies. Sci. Transl. Med. 3, 108ps44 (2011).
Thor Straten, P. et al. In situ T cell responses against melanoma comprise high numbers of locally expanded T cell clonotypes. J. Immunol. 163, 443–447 (1999).
Kobayashi, E. et al. A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nat. Med. 19, 1542–1546 (2013).
Cohen, C. J. et al. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 67, 3898–3903 (2007).
Stanke, J. et al. A flow cytometry-based assay to assess minute frequencies of CD8+ T cells by their cytolytic function. J. Immunol. Methods 360, 56–65 (2010).
Santegoets, S. J. et al. Monitoring regulatory T cells in clinical samples: consensus on an essential marker set and gating strategy for regulatory T cell analysis by flow cytometry. Cancer Immunol. Immunother. 64, 1271–1286 (2015).
Acknowledgements
We thank all patients and their families for participating in this clinical trial; the Data Safety Monitoring Board (J. Rich, O. Chinot and C. Huber); S. Hess, M. Krauss (FGK) and S. Becker (spm2) for their contributions to trial management; I. Groskopf and M. Adis for technical support; T. Hillemann, M. Erdeljan and L. Alten for contributions to the revisions. A. Salazar (Oncovir) provided poly-ICLC. GAPVAC was supported by the European Union’s FP7-HEALTH Research and Innovation funding program (number 305061). Further support information can be found in Supplementary Notes.
Reviewer information
Nature thanks E. Jaffee, M. Lim and C. Melief for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
C.M.B., C.H., H.-G.R., H.O., H.S.-J., J.C.C., N.H., O.S., S.K.-C., S.H., T.W., U.S. and W.W. developed the GAPVAC concept and trial design. D. Migliorini, F.M.-R., G.T., H.S.P., J.R.K., J.R., J.S., M. Skardelly, P.-Y.D., U.L. and W.W. recruited and treated patients. A.K.-B., B.R., C.R., J.L., K.F., M.M., N.H., N.P., P.-Y.D. and W.W overviewed the clinical trial conduct and assured compliance with Good Clinical Practice (GCP). A.v.D., A.D.T., C.S., D.C., J.C.C., J.F., K.F., M.L., N.H., O.S., S.H., S.K., S.K.-C., T.W. and V.B. contributed and/or evaluated data for the APVAC vaccines. B.P., H.-G.R., J.P., M. Stieglbauer, S.D. and S.S. contributed to APVAC manufacturing. A.U., C.G., C.W., D. Maurer, E.D., E.G., E.R., L.B., M.P., M.J.P.W., R.M., V.D. and W.W. contributed to immune response analyses. C.G., E.R., M.I., M.J.P.W., P.t.S., S.H.v.d.B. and V.D. contributed to the TIL analyses and together with C.M.B., G.T., M. Skardelly, J.R.K., H.S.P., P.-Y.D. and D. Migliorini contributed to the TIL protocol. A.A., A.U., B.S., C.H.O., K.M., K.F., R.M., U.S. and V.B. contributed further biomarker data. J.F., J.L., F.H., K.K., N.H., P.-Y.D., S.K.-C. and W.W. evaluated the clinical data. A.U., C.G., C.W., H.O., J.L., K.K., N.H., P.-Y.D., R.M., S.H.v.d.B., S.K.-C., V.D. and W.W. wrote the manuscript. A.D.T., C.S., J.F., K.F., M.L., N.H., O.S., S.H., S.K., S.K.-C., T.W. and V.B. contributed to Extended Data Fig. 1. A.U., C.W., D. Maurer and R.M. contributed to Fig. 2a–c, f, 3, 4 and Extended Data Figs. 1d, 3, 4a–c, 5, 6, 7b and 10. C.G., E.R. and H.-G.R. contributed to Fig. 5c, d and Extended Data Fig. 7a. V.D. and P.-Y.D. contributed to Extended Data Fig. 3d. M.P., L.B., E.G. and W.W. contributed to Fig. 2d, e and Extended Data Fig. 3e–g. K.M., A.U., R.M. and C.H.O. contributed to the Treg cell data in Extended Data Fig. 4a, e, f. J.L., F.H., K.K., N.H., P.-Y.D. and W.W. contributed to Fig. 5a, Table 1, Extended Data Figs. 2, 9a, b and Supplementary Table 5. K.F., V.B. and U.S. contributed to Fig. 5b. C.G., E.R., M.I., M.J.P.W., P.t.S., S.H.v.d.B. and V.D. contributed to Extended Data Fig. 8.
Corresponding author
Ethics declarations
Competing interests
N.H., S.K.-C., A.U., R.M., K.K., C.W., J.L., O.S., J.F., M.M., N.P., S.D., F.H., D. Maurer, T.W., C.R. and H.S.-J. are employees of Immatics Biotechnologies. C.S. and B.R. contributed as former employees of Immatics to this work. T.W., C.R. and H.S.-J. are shareholders of Immatics. U.S., E.D., S.H., A.K.-B., K.F. and V.B. work for BioNTech. U.S. is CEO, co-founder and shareholder of BioNTech and inventor on patents related to personalized cancer immunotherapy. C.H. is co-founder, shareholder, advisor and member of the supervisory board of BioNTech AG, co-founder, advisor and member of the supervisory board of TRON AG. He also received honoraria from Immatics, Affiris, Abivax, Apceth, Suppremol and Vaxxim as member of advisory boards. M.L. works as a consultant and receives research funding from BioNTech and is co-inventor on patents related to the personalized cancer vaccine. A.D.T. is co-inventor on patents related to the personalized cancer vaccine. S.K. is vice president and advisor for BioNTech. C.M.B. contributed as former employee of BioNTech to this work. J.C.C. worked as an advisor for BioNTech. B.P. and J.P. are employees of BCN Peptides. H.-G.R. has received research grants from Boehringer-Ingelheim as well as honoraria from CureVac for advisory board participation and consulting for Navigo Proteins, AstraZeneca, MSD, BMS and MedImmune. C.G. has received research grants from Boehringer-Ingelheim and from RhoVac ApS. M.P. has received research grants from Bayer, Pfizer and Novartis as well as honoraria for lectures, advisory board participation or consulting from Bayer, Merck, Novartis, Roche, Affiris and Medac. G.T. has received research grants from Roche Diagnostics as well as honoraria for lectures, advisory board participation or consulting from Medac, BMS, Novocure and AbbVie. S.H.v.d.B. has received research grants from ISA Pharmaceuticals, Kite Pharma, Innate Pharma and IO-Biotech as well as honoraria for advisory board participation or consulting from ISA Pharmaceuticals, IO-Biotech, PCI Biotech, and GSK. P.t.S. has received honoraria from RhoVac. J.R. has received research grants from Bayer and Novartis as well as honoraria for lectures, advisory board participation or consulting from Novartis, Eli Lilly, Orion Pharmaceuticals, Servier Pharmaceuticals, Peptomyc, Merck Sharp & Dohme, Kelun Pharmaceutical/Klus Pharma and Spectrum Pharmaceuticals. H.O. has received research grants from Agios Pharmaceuticals, Ono Pharmaceutical as well as honoraria for lectures, advisory board participation or consulting from Bristol-Myers Squibb, Alexion Pharmaceuticals, Amal Therapeutics, Agios Pharmaceuticals and Eureka Therapeutics. J.R.K. has received research grants from Amgen, Astra Zeneca, Novartis, Sanofi and Philips as well as honoraria for lectures, advisory board participation or consulting from Amgen, Astra Zeneca, Eisai, Novartis, Pfizer, Roche and Tesaro. P.-Y.D. has received research grants from Immatics as well as honoraria for lectures, advisory board participation or consulting from BMS. W.W. has received research support from Apogenix, Boehringer Ingelheim, Pfizer, Roche and Vaximm. S.S., V.D., U.L., M.I., F.M.-R., C.H.O, H.S.P., A.v.D., J.S., M. Skardelly, D. Migliorini, A.A., B.S., E.R., M. Stieglbauer, D.C., L.B., E.G., K.M. and M.J.P.W. have no conflicts to disclose.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
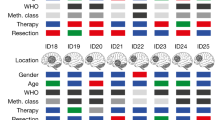
Extended Data Fig. 1 APVAC1 and APVAC2 processes lead to truly personalized vaccine formulations.
a, Summary of biomarker data used to define APVAC1 for patient 5. Immunogenicity was calculated as the percentage of peptide-reactive wells (n = 12) with patient-derived PBMCs. Presentation of peptides on the tumour of patient 5 is shown as detected by mass spectrometry (present or absent). Exclusivity indicates the presence or absence of peptides in normal tissue samples (n = 394). Individual overpresentation ratio at the level of the HLA peptidome on comparing the tumour of patient 5 and the average of normal tissues or the limit of detection for the respective peptide (if it was never found in normal tissues). The mRNA overexpression ratio of the source genes in the tumour of patient 5 compared to the average of normal tissues. n.d., no data; genes not covered by the microarray that was used. Data were integrated into a rank score as basis for selection into APVAC1 (see Methods for details). b, Peptide presentation of PTP-013 on normal tissues (n = 394, not detected), glioblastoma specimens (identified on 7 out of 33 independent samples, median indicated), and the tumour of patient 5 as analysed by mass spectrometry. For patient 5, mean signal and signal range of n = 5 replicate mass spectrometry runs are shown. c, mRNA expression of PTPRZ1—which encodes PTP-013—analysed by mRNA microarray analysis in samples of pooled normal tissue mRNA, glioblastoma tumours (n = 26, median indicated) and tumour of patient 5. n.d., no data. b, c, BLV, blood vessels; BRA, brain; DIG, digestive glands; FRP, female reproductive organs; HRT, heart; HEM, haematopoietic cells; HOG, hormonal glands; INT, intestine; KID, kidney; LIV, liver; LYM, lymphatic tissue; MRP, male reproductive organs; NER, nervous system; RES, respiratory; SKM, skeletal muscle; SKI, skin; STO, stomach/oesophagus; URI, urinary system; OTH, other. d, Immunogenicity pretesting data for PTP-013 in pretreatment PBMCs of patient 5. Cells (1 × 106 per well, n = 12 wells tested) were stimulated with each A*02 warehouse peptide separately. After in vitro stimulation, wells were analysed for peptide-specific CD8+ T cells using 2D multimer staining. Representative positive (7 out of 12) and negative (5 out of 12) wells are shown. e, Number of selections into APVAC1 compositions for the 33 A*02 warehouse peptides (n = 16 patients). f, APVAC2 selection process for mutated neoepitopes. g, The four APVAC2 candidates entering test synthesis for patient 11 are shown (patient dropped out before being vaccinated with APVAC2). Best predicted HLA class I epitopes are underlined. Red, mutated position. h, APVAC2 composition overview for GAPVAC-101 patients vaccinated with APVAC2 (n = 11).
Extended Data Fig. 2 APVAC composition, vaccination schedule, exposure to drug and standard therapy, and course of patients.
a, Composition of personalized APVAC1 and APVAC2 drug products. b, Vaccination schedule for APVAC1 and APVAC2 and the immunomodulators. Note that APVAC1 and APVAC2 vaccinations started independently on day 15 of the first and fourth adjuvant TMZ cycle, respectively. In the default schedule, both APVACs were co-applied at the ninth APVAC1 and fourth APVAC2 vaccination, and at all four-weekly applications following the tenth APVAC1 and seventh APVAC2 vaccination. In case of delayed or failed APVAC production, alternative schedules were in place to optimally align vaccinations with standard therapy (data not shown). Any assessments at a vaccination visit, including blood draws for PBMC isolation, were performed prior to vaccinations. c, Patient exposure to study drugs. d, Exposure to standard therapy. Statistics on number of applications, dose and time lines between therapy phases are provided for all enrolled GAPVAC-101 patients (n = 16). SD, standard deviation. Reference values for the standard therapy are provided29. *Completed treatment assumed with 75 mg m−2 for 6–7 weeks calculated with 1.75-m2 body surface area. **According to treatment schedule, patient data not published29. e, CONSORT-like diagram for patients in the GAPVAC-101 trial. Completion for APVAC1 and APVAC2 vaccinations according to the trial protocol equals to at least 11 and 8 vaccinations, respectively. Three patients left the study before receiving APVAC2: two patients withdrew their consents; one patient experienced a grade 3 seizure (unrelated to vaccinations) and withdrew from the study. Additionally, no biomarker data for definition of APVAC2 was available for patient 4, because the tumour specimen was necrotic.
Extended Data Fig. 3 Functional characterization of APVAC responses.
a, Expression of activation/exhaustion marker PD-1 for three different immune responses pre-vaccination and at different post-vaccination time points. Top, example response to viral marker peptide HBV-001. Middle and bottom, example APVAC1-specific CD8+ T cell responses. Numbers in plots show ratio of PD-1 mean fluorescence intensity (MFI) for target-specific cells (blue curve) divided by MFI of total CD8+ T cells (grey curve). b, Summary of PD-1 evaluations as shown in a. Maximum MFI ratio for each CD8+ T cell response to APVAC1 peptides (n = 16) and to the viral marker peptide HBV-001 (n = 2) from four patients (2, 5, 8 and 14). Data are mean and individual values. c, Flow cytometry data for Vital-FR assay shown in Fig. 2c. PTP-013-specific CD8+ T cells were sorted from post-vaccination T cells from patient 16, in vitro-expanded for three weeks and tested for killing of CFSE-labelled K562-A2 cells loaded with titrated amounts of PTP-013 peptide in comparison to FarRed-labelled K562-A2 cells loaded with an irrelevant peptide. Data at the four lowest peptide concentrations are shown. Owing to limited availability of patient PBMCs, this experiment was performed only once with n = 3 replicates. d, BCA-002-reactive T cells from pre-vaccination PBMCs from patient 11 were cloned and expanded with PHA and IL-2. Obtained clones were tested in a flow-cytometry-based cytotoxicity assay using T2 cells loaded with titrated amounts of peptide. Results for n = 10 different clones are shown with calculated EC50 values between 49 pM and 2.8 nM (means of 2–3 replicates per clone). e, NLGN4X T cells as described for Fig. 2e were able to specifically lyse the A*02+ glioblastoma cell line P3XX that endogenously expresses NLGN4X (LDH release assay). HD T cells, healthy donor-derived T cells that are not target-specific. Results from n = 3 replicates are shown (median and range). f, NLGN4X T cells were either co-cultured with the glioblastoma cell line U87MG transduced with NLGN4X–nLuc (red), irrelevant control NRCAM–nLuc (black) or in the absence of target cells (blue). With increasing E:T ratio, complete eradication of NLGN4X–nLuc-expressing target cells is shown (NanoLuc luciferase-based killing assays, median and range from n = 4 replicates shown). RLU, relative luminometer units. Additional functional data of warehouse peptides has previously been published18. g, Expression of the source genes PTPRZ1 and NLGN4X in the used glioblastoma cell lines compared to the housekeeping gene ACTB (which encodes β-actin) measured by quantitative PCR. Boxes describe range and median from n = 4–6 replicates.
Extended Data Fig. 4 Additional immune response data.
a, APVAC1 CD8+ T cell response parameters including MCIF (see Methods), frequencies of Treg cells at baseline and predominant phenotypes of APVAC-induced CD4+ T cell responses for APVAC1 immune-evaluable patients (n = 13). n.e., not evaluable. b, Frequency of NLGN4X-001-specific T cells of patient 2 by differentiation phenotype summarized from the flow cytometry data shown in Fig. 2b. c, TH1 CD4+ T cell response against the APVAC1 HLA class II peptide PTP-010 in PBMCs from patient 8, confirming ELISPOT results (Fig. 5d) and reactivity in TILs (Fig. 5c). d, MCIF for GAPVAC-101 patients (evaluable for n = 13 patients) by APVAC1-specific CD4+ T cell response type. For response type colour codes, see a. e, Pretreatment Treg cells of patients with high or low MCIF (evaluable for n = 12 patients). f, Peak frequencies of IFNγ-producing APVAC1 pan-DR peptide-specific CD4+ T cells in patients with high frequencies of Treg cells (>median: 3.6%) versus those with low frequencies of Treg cells. Each dot represents one response (n = 24, two for each of the 12 evaluable patients). d–f, Median and 95% confidence interval of the median are depicted, P values determined by two-sided Mann–Whitney U-test. g, Immune responder rates (with and without memory shift) in GAPVAC-101 and a previous vaccine trial (IMA950-101) with the invariant multi-peptide vaccine IMA95016 to the warehouse antigens BCA-002, NLGN4X-001, PTP-005 and the viral marker HBV-001 shared by both vaccines. Antigen responses were evaluated in both cases by the same ex vivo multimer 2D assay. Numbers below bars indicate the number of patients analysed for the respective antigen. In IMA950-101 with a comparable population of patients with glioblastoma, specific CD8+ T cell responses were rarely detected ex vivo. In the GAPVAC-101 trial, measurable and sustained CD8+ memory T cell responses (with at least duplication of memory cell frequencies) against the same tumour antigens were frequent. The pre-immunogenicity testing in GAPVAC-101 is probably not the main cause of the observed higher response rate, as progenitor T cells for the three compared antigens are abundant in A*02+ individuals (see Supplementary Table 1). h, APVAC2-specific CD4+ T cell responses for all evaluable patients (n = 10). Cell frequencies are only shown for key cytokine-positive T cells. P, prior to APVAC2 vaccination; 1–4, post-treatment measurements. Phenotype assignments as indicated. Additional CD8+ T cell responses to APVAC2 peptides are indicated with the detected lead cytokine. *Pre-defined response criteria not met. #Response to 13-M06 was negative for any key cytokine. TNF+ cells are shown. a–h, Owing to limited availability of patient PBMCs, immune response analyses were only performed once per patient. For assays with limited patient materials, see ‘Statistical analyses’ in Methods.
Extended Data Fig. 5 Ex vivo ICS flow cytometry data for APVAC1-induced CD4+ T cell responses.
a, Example flow cytometry data for responses shown in Fig. 2f (CD4+ T cell response of patient 7 to pan-DR antigen BIR-002). IL-5 was negative at all time points (data not shown). b, Mock control stimulation for a. a, b, Owing to limited availability of PBMCs, immune response analyses were performed only once per patient. For assays with limited patient materials, see ‘Statistical analyses’ in Methods.
Extended Data Fig. 6 Pan-ICS flow cytometry data.
a, Pan-ICS flow cytometry data for CD4+ T cell response of patient 16 to mutated APVAC2 peptide 16-M06 as summarized in Fig. 4a. Production of indicated cytokines pretreatment (Pre) and at three different post-treatment pools (Post 1 to Post 3) is shown. Post 2 and Post 3 pools originate from the continued vaccination phase (see Fig. 1). Dot plots are gated on CD8− lymphocytes. Note that the pan-ICS cytokine panel differs from the one used for the ex vivo ICS assay (see Methods). b, Mock control stimulation for a. c, Pan-ICS flow cytometry data for CD8+ T cell response of patient 15 against the mutated APVAC2 peptide 15-M05 as summarized in Fig. 4b. A pool of all possible peptides that were nine amino acids long from 15-M05 was used for read-out. Production of indicated cytokines is shown pretreatment (Pre) and at two different post-treatment pools (Post 1 and Post 2). Dot plots are gated on CD4− lymphocytes. Note that the APVAC2 analysis time points are not identical to APVAC1 analysis time points owing to the independent start of vaccination (Fig. 1). d, Mock control stimulation for c. a–d, Owing to limited availability of PBMCs, immune response analyses were only performed once per patient. For assays with limited patient materials, see ‘Statistical analyses’ in Methods.
Extended Data Fig. 7 Additional data on APVAC2-induced, neoepitope-specific immune responses.
a, Cross-reactivity of two neoepitope-directed, APVAC2-induced CD4+ T cell responses with the corresponding wild-type peptides was assessed by ICS. Top, response for 08-M01 was specific to the mutated peptide, whereas the response for 16-M06 (bottom) showed considerable cross-reactivity with the wild-type peptide. Owing to limited availability of patient PBMCs, this experiment was performed only once. b, Ex vivo 2D multimer data for CD8+ T cell response of patient 14 against the predicted neoepitope HMKVSVYLL contained in the vaccinated peptide 14-M09 as summarized in Fig. 4c. Note that this epitope did not show the highest ranking during APVAC2 selection (underlined in Supplementary Table 4). The pretreatment pool and one post-treatment pool were evaluated. Top, 2D multimer staining. Numbers in plots indicate the frequency of specific cells among total CD8+ T cells. Middle, differentiation phenotyping of specific T cells (blue dots) and total CD8+ T cells (grey dots). Numbers in plots indicate the percentage of specific CD8+ T cells with indicated phenotype (colour code as in Fig. 2a, b). Bottom, PD-1 expression (blue curve, specific CD8+ T cells; grey dots, all CD8+ T cells).
Extended Data Fig. 8 TIL isolation and expansion in glioblastoma.
A harmonized TIL isolation and expansion protocol for glioblastoma specimens was developed in the preclinical phase of the GAPVAC project (see Methods). a, Cells were expanded in the presence of recombinant human IL-2 at three concentrations. One centre also performed expansion of melanoma and oropharyngeal TILs for comparison. The number of independent tumours compared across laboratories (preclinical samples) are shown in brackets. Data are mean, s.e.m. and individual values for each cell subset. b, Expansion of glioma TILs in the presence or absence of the anti-CD3 antibody OKT3. Indicated number of pieces of tumour obtained from three patients with glioblastoma (outside the GAPVAC-101 trial) were cut in fragments and transferred into 24-well plates with one fragment per well. Fragments were cultured in the absence or presence of 30 ng ml−1 anti-CD3 antibody (OKT3). Expanded cells were collected at the indicated days and subjected to phenotyping by flow cytometry. Percentages of total CD3+, CD4+ and CD8+ T cells and total cell yield are indicated. c, Compositions of TIL cultures derived at one centre from four different patients (6, 7, 8 and 9) included in the GAPVAC-101 trial. Each dot represents one stain performed either on independent culture wells or after pooling several wells (numbers of independent stainings per patient are indicated). The glioblastoma-derived TILs generally comprised a mixture of CD4+ and CD8+ T cells that varied between individual cultures. Bars indicate the median over all cultures. d, Example of TCR clonotype mapping used for a comparative analysis of clonotypes detected ex vivo in the fresh tumour relapse tissue (FT) and in n = 3 independent TIL cultures of patient 8. Identical DNA sequences resolve at identical positions in the gel. One prominent BV14 clonotype present in the fresh tissue (arrow) is not amplified in the TIL cultures (left), whereas two of several BV21 clonotypes in the fresh tumour (*) are also detected in the TILs (right). At least n = 8 clonotypes per matched set of samples were compared.
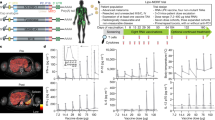
Extended Data Fig. 9 Survival data for GAPVAC-101 patients.
a, b, Overall survival (OS, a) and progression-free survival (PFS, b) from diagnosis for all patients that received at least one APVAC vaccination (n = 15). Kaplan–Meier estimates of median survival are provided. c, Regulatory pathway for active personalized vaccines for GAPVAC-101 compared to conventional immunotherapies. The pathway for clinical trial application (CTA) for GAPVAC-101 was pre-discussed in scientific advice and pre-Investigational New Drug meetings with the German Paul-Ehrlich-Institute (PEI) and the US Food and Drug Administration (FDA), respectively. Key characteristics in the trial approval process were: (1) standardization of a drug composition process based on highly variable, patient-individual biomarker data instead of a fixed drug composition; (2) development of a GMP-compliant core process for manufacturing of variable, personalized multi-peptide compositions instead of an invariable manufacturing process; (3) clinical trial application based on exemplary data from representative preclinical drug substance batches; and (4) provision of certificates of analysis for all APVACs to the authorities on a continuous basis during the trial (twice yearly).
Extended Data Fig. 10 Gating strategies for flow cytometry assays.
a, Gating strategy and Boolean gating for ex vivo 2D multimer assay (CD8+ T cell immune response analysis, APVAC1 and APVAC2). b, Gating strategy for ex vivo class II ICS assay (CD4+ T cell immune response analysis, APVAC1). c, Gating strategy for HLA class I and class II pan-ICS assay (CD4+ and CD8+ T cell immune response analysis, APVAC2). d, Gating strategy for Treg cell assay.
Supplementary information
Supplementary Information
This file contains Supplementary Methods sections 1 and 2, Supplementary Notes and Supplementary Tables 1, 2, 4 and 5.
Supplementary Table 3
A list of non-synonymous mutations in the tumours of GAPVAC-101 patients.
Rights and permissions
About this article
Cite this article
Hilf, N., Kuttruff-Coqui, S., Frenzel, K. et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565, 240–245 (2019). https://doi.org/10.1038/s41586-018-0810-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0810-y
This article is cited by
-
Revealing the role of SPP1+ macrophages in glioma prognosis and therapeutic targeting by investigating tumor-associated macrophage landscape in grade 2 and 3 gliomas
Cell & Bioscience (2024)
-
mRNA-based precision targeting of neoantigens and tumor-associated antigens in malignant brain tumors
Genome Medicine (2024)
-
Challenges in developing personalized neoantigen cancer vaccines
Nature Reviews Immunology (2024)
-
T-cell stimulating vaccines empower CD3 bispecific antibody therapy in solid tumors
Nature Communications (2024)
-
Computational immunogenomic approaches to predict response to cancer immunotherapies
Nature Reviews Clinical Oncology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.