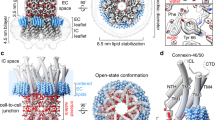
Abstract
Gap junctions establish direct pathways for cell-to-cell communication through the assembly of twelve connexin subunits that form intercellular channels connecting neighbouring cells. Co-assembly of different connexin isoforms produces channels with unique properties and enables communication across cell types. Here we used single-particle cryo-electron microscopy to investigate the structural basis of connexin co-assembly in native lens gap junction channels composed of connexin 46 and connexin 50 (Cx46/50). We provide the first comparative analysis to connexin 26 (Cx26), which—together with computational studies—elucidates key energetic features governing gap junction permselectivity. Cx46/50 adopts an open-state conformation that is distinct from the Cx26 crystal structure, yet it appears to be stabilized by a conserved set of hydrophobic anchoring residues. ‘Hot spots’ of genetic mutations linked to hereditary cataract formation map to the core structural–functional elements identified in Cx46/50, suggesting explanations for many of the disease-causing effects.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession number EMD-9116. Coordinates for Cx46 and Cx50 atomic models have been deposited in the Protein Data Bank under accession codes 6MHQ and 6MHY. The original multi-frame micrographs have been deposited in the Electron Microscopy Public Image Archive under accession code EMPIAR-10212.
References
Goodenough, D. A. & Paul, D. L. Gap junctions. Cold Spring Harb. Perspect. Biol. 1, a002576 (2009).
Delmar, M. et al. Connexins and disease. Cold Spring Harb. Perspect. Biol. 10, a029348 (2018).
Garcia, I. E. et al. Connexinopathies: a structural and functional glimpse. BMC Cell Biol. 17 (Suppl 1), 17 (2016).
Aasen, T., Mesnil, M., Naus, C. C., Lampe, P. D. & Laird, D. W. Gap junctions and cancer: communicating for 50 years. Nat. Rev. Cancer 16, 775–788 (2016).
Sosinsky, G. E. & Nicholson, B. J. Structural organization of gap junction channels. Biochim. Biophys. Acta 1711, 99–125 (2005).
Sohl, G. & Willecke, K. Gap junctions and the connexin protein family. Cardiovasc. Res. 62, 228–232 (2004).
Cottrell, G. T. & Burt, J. M. Functional consequences of heterogeneous gap junction channel formation and its influence in health and disease. Biochim. Biophys. Acta 1711, 126–141 (2005).
Beyer, E. C. & Berthoud, V. M. Gap junction structure: unraveled, but not fully revealed. F1000 Res. 6, 568 (2017).
Grosely, R. & Sorgen, P. L. A history of gap junction structure: hexagonal arrays to atomic resolution. Cell Commun. Adhes. 20, 11–20 (2013).
Maeda, S. et al. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 (2009).
Bennett, B. C. et al. An electrostatic mechanism for Ca2+-mediated regulation of gap junction channels. Nat. Commun. 7, 8770 (2016).
Mathias, R. T., White, T. W. & Gong, X. Lens gap junctions in growth, differentiation, and homeostasis. Physiol. Rev. 90, 179–206 (2010).
Konig, N. & Zampighi, G. A. Purification of bovine lens cell-to-cell channels composed of connexin44 and connexin50. J. Cell Sci. 108, 3091–3098 (1995).
Jiang, J. X. & Goodenough, D. A. Heteromeric connexons in lens gap junction channels. Proc. Natl Acad. Sci. USA 93, 1287–1291 (1996).
Shearer, D., Ens, W., Standing, K. & Valdimarsson, G. Posttranslational modifications in lens fiber connexins identified by off-line-HPLC MALDI-quadrupole time-of-flight mass spectrometry. Invest. Ophthalmol. Vis. Sci. 49, 1553–1562 (2008).
Wang, Z. & Schey, K. L. Phosphorylation and truncation sites of bovine lens connexin 46 and connexin 50. Exp. Eye Res. 89, 898–904 (2009).
Kwon, T. et al. Molecular dynamics simulations of the Cx26 hemichannel: insights into voltage-dependent loop-gating. Biophys. J. 102, 1341–1351 (2012).
Bai, D. Structural analysis of key gap junction domains–lessons from genome data and disease-linked mutants. Semin. Cell Dev. Biol. 50, 74–82 (2016).
Koval, M., Molina, S. A. & Burt, J. M. Mix and match: investigating heteromeric and heterotypic gap junction channels in model systems and native tissues. FEBS Lett. 588, 1193–1204 (2014).
Unger, V. M., Kumar, N. M., Gilula, N. B. & Yeager, M. Three-dimensional structure of a recombinant gap junction membrane channel. Science 283, 1176–1180 (1999).
Fleishman, S. J., Unger, V. M., Yeager, M. & Ben-Tal, N. A Cα model for the transmembrane alpha helices of gap junction intercellular channels. Mol. Cell 15, 879–888 (2004).
Gong, X. Q. & Nicholson, B. J. Size selectivity between gap junction channels composed of different connexins. Cell Commun. Adhes. 8, 187–192 (2001).
Goldberg, G. S., Valiunas, V. & Brink, P. R. Selective permeability of gap junction channels. Biochim. Biophys. Acta 1662, 96–101 (2004).
Varland, S., Osberg, C. & Arnesen, T. N-terminal modifications of cellular proteins: the enzymes involved, their substrate specificities and biological effects. Proteomics 15, 2385–2401 (2015).
Berneche, S. & Roux, B. Energetics of ion conduction through the K+ channel. Nature 414, 73–77 (2001).
Corry, B. & Thomas, M. Mechanism of ion permeation and selectivity in a voltage gated sodium channel. J. Am. Chem. Soc. 134, 1840–1846 (2012).
Ulmschneider, M. B. et al. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc. Natl Acad. Sci. USA 110, 6364–6369 (2013).
Srinivas, M. et al. Voltage dependence of macroscopic and unitary currents of gap junction channels formed by mouse connexin50 expressed in rat neuroblastoma cells. J. Physiol. 517, 673–689 (1999).
Hopperstad, M. G., Srinivas, M. & Spray, D. C. Properties of gap junction channels formed by Cx46 alone and in combination with Cx50. Biophys. J. 79, 1954–1966 (2000).
Oh, S., Rubin, J. B., Bennett, M. V., Verselis, V. K. & Bargiello, T. A. Molecular determinants of electrical rectification of single channel conductance in gap junctions formed by connexins 26 and 32. J. Gen. Physiol. 114, 339–364 (1999).
Tong, X. et al. The first extracellular domain plays an important role in unitary channel conductance of Cx50 gap junction channels. PLoS ONE 10, e0143876 (2015).
Trexler, E. B., Bukauskas, F. F., Kronengold, J., Bargiello, T. A. & Verselis, V. K. The first extracellular loop domain is a major determinant of charge selectivity in connexin46 channels. Biophys. J. 79, 3036–3051 (2000).
Oh, S., Verselis, V. K. & Bargiello, T. A. Charges dispersed over the permeation pathway determine the charge selectivity and conductance of a Cx32 chimeric hemichannel. J. Physiol. 586, 2445–2461 (2008).
Kronengold, J., Trexler, E. B., Bukauskas, F. F., Bargiello, T. A. & Verselis, V. K. Pore-lining residues identified by single channel SCAM studies in Cx46 hemichannels. Cell Commun. Adhes. 10, 193–199 (2003).
Zonta, F., Polles, G., Zanotti, G. & Mammano, F. Permeation pathway of homomeric connexin 26 and connexin 30 channels investigated by molecular dynamics. J. Biomol. Struct. Dyn. 29, 985–998 (2012).
Lopez, W. et al. Mechanism of gating by calcium in connexin hemichannels. Proc. Natl Acad. Sci. USA 113, E7986–E7995 (2016).
Harris, A. L. & Contreras, J. E. Motifs in the permeation pathway of connexin channels mediate voltage and Ca2+ sensing. Front. Physiol. 5, 113 (2014).
Srinivas, M., Calderon, D. P., Kronengold, J. & Verselis, V. K. Regulation of connexin hemichannels by monovalent cations. J. Gen. Physiol. 127, 67–75 (2006).
Trexler, E. B., Bennett, M. V., Bargiello, T. A. & Verselis, V. K. Voltage gating and permeation in a gap junction hemichannel. Proc. Natl Acad. Sci. USA 93, 5836–5841 (1996).
Suchyna, T. M. et al. Different ionic selectivities for connexins 26 and 32 produce rectifying gap junction channels. Biophys. J. 77, 2968–2987 (1999).
Veenstra, R. D. Size and selectivity of gap junction channels formed from different connexins. J. Bioenerg. Biomembr. 28, 327–337 (1996).
Veenstra, R. D. et al. Selectivity of connexin-specific gap junctions does not correlate with channel conductance. Circ. Res. 77, 1156–1165 (1995).
Nicholson, B. J. et al. The molecular basis of selective permeability of connexins is complex and includes both size and charge. Braz. J. Med. Biol. Res. 33, 369–378 (2000).
Ebihara, L., Xu, X., Oberti, C., Beyer, E. C. & Berthoud, V. M. Co-expression of lens fiber connexins modifies hemi-gap-junctional channel behavior. Biophys. J. 76, 198–206 (1999).
Xin, L. & Bai, D. Functional roles of the amino terminal domain in determining biophysical properties of Cx50 gap junction channels. Front. Physiol. 4, 373 (2013).
Luo, Y., Rossi, A. R. & Harris, A. L. Computational studies of molecular permeation through connexin26 channels. Biophys. J. 110, 584–599 (2016).
Kwon, T., Harris, A. L., Rossi, A. & Bargiello, T. A. Molecular dynamics simulations of the Cx26 hemichannel: evaluation of structural models with Brownian dynamics. J. Gen. Physiol. 138, 475–493 (2011).
Verselis, V. K., Ginter, C. S. & Bargiello, T. A. Opposite voltage gating polarities of two closely related connexins. Nature 368, 348–351 (1994).
Peracchia, C. & Peracchia, L. L. Inversion of both gating polarity and CO2 sensitivity of voltage gating with D3N mutation of Cx50. Am. J. Physiol. Cell Physiol. 288, C1381–C1389 (2005).
Srinivas, M., Kronengold, J., Bukauskas, F. F., Bargiello, T. A. & Verselis, V. K. Correlative studies of gating in Cx46 and Cx50 hemichannels and gap junction channels. Biophys. J. 88, 1725–1739 (2005).
Xin, L., Nakagawa, S., Tsukihara, T. & Bai, D. Aspartic acid residue D3 critically determines Cx50 gap junction channel transjunctional voltage-dependent gating and unitary conductance. Biophys. J. 102, 1022–1031 (2012).
Beyer, E. C., Ebihara, L. & Berthoud, V. M. Connexin mutants and cataracts. Front. Pharmacol. 4, 43 (2013).
Pascolini, D. & Mariotti, S. P. Global estimates of visual impairment: 2010. Br. J. Ophthalmol. 96, 614–618 (2012).
Shiels, A. & Hejtmancik, J. F. Mutations and mechanisms in congenital and age-related cataracts. Exp. Eye Res. 156, 95–102 (2017).
Kistler, J., Christie, D. & Bullivant, S. Homologies between gap junction proteins in lens, heart and liver. Nature 331, 721–723 (1988).
Kistler, J., Schaller, J. & Sigrist, H. MP38 contains the membrane-embedded domain of the lens fiber gap junction protein MP70. J. Biol. Chem. 265, 13357–13361 (1990).
White, T. W., Bruzzone, R., Goodenough, D. A. & Paul, D. L. Mouse Cx50, a functional member of the connexin family of gap junction proteins, is the lens fiber protein MP70. Mol. Biol. Cell 3, 711–720 (1992).
Reichow, S. L. et al. Allosteric mechanism of water-channel gating by Ca2+-calmodulin. Nat. Struct. Mol. Biol. 20, 1085–1092 (2013).
Gold, M. G. et al. AKAP2 anchors PKA with aquaporin-0 to support ocular lens transparency. EMBO Mol. Med. 4, 15–26 (2012).
Reichow, S. L. & Gonen, T. Noncanonical binding of calmodulin to aquaporin-0: implications for channel regulation. Structure 16, 1389–1398 (2008).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protocols 1, 2856–2860 (2006).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Myers, J. B. et al. The CaMKII holoenzyme structure in activation-competent conformations. Nat. Commun. 8, 15742 (2017).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–38 (1996).
Locke, D., Bian, S., Li, H. & Harris, A. L. Post-translational modifications of connexin26 revealed by mass spectrometry. Biochem. J. 424, 385–398 (2009).
Grubmuller, H., Heymann, B. & Tavan, P. Ligand binding: molecular mechanics calculation of the streptavidin–biotin rupture force. Science 271, 997–999 (1996).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles J. Chem. Theory Comput. 8, 3257–3273 (2012).
Zuckerman, D. M. Statistical Physics of Biomolecules: An Introduction, 1st edn (CRC, Boca Raton, 2010).
Im, W., Seefeld, S. & Roux, B. A grand canonical Monte Carlo–Brownian dynamics algorithm for simulating ion channels. Biophys. J. 79, 788–801 (2000).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We thank T. Gonen for early support of this work; T. White, L. David, U. Adhikari and B. Mostofian for helpful discussions; the staff at the OHSU Multiscale Microscopy Core and Advanced Computing Center, and W. Garrick (PSU) for their assistance and training. C.V.R. and D.S.C. are supported by funding from the European Research Council (No. 695511-ENABLE). D.M.Z. and C.C.Y. are supported by the Center for Spatial Systems Biomedicine at OHSU. S.L.R. is supported by the Medical Research Foundation of Oregon and the National Institutes of Health (R35-GM124779).
Author information
Authors and Affiliations
Contributions
J.B.M. and B.G.H. contributed equally. J.B.M. and C.C.Y. collected the cryo-EM datasets. J.B.M. performed image processing and atomic modelling of Cx46/50. S.E.O. performed protein purification and negative-stain electron microscopy studies. B.G.H. conducted and analysed the molecular dynamics simulations. D.S.C conducted the cross-linking studies and MS/MS analysis. C.V.R. contributed to the experimental design of MS/MS studies. D.M.Z. contributed to the experimental design and statistical analysis of the molecular dynamics simulations. All authors contributed to manuscript preparation. S.L.R. provided overall guidance to the design and execution of this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Preliminary structural analysis of native lens Cx46/50 gap junction intercellular channels.
a, Denaturing protein gel and western blot analysis of purified Cx46/50 (also known as MP38) isolated from lens core tissue. Protein bands corresponding to Cx46 and Cx50 co-migrate at a molecular weight of ~38 kDa, consistent with age-dependent proteolytic cleavage within the C-terminal domain of both isoforms16. Experiment performed 3 times with similar results. b, SEC elution profile of Cx46/50 gap junctions reconstituted in decyl-maltoside (DM, grey trace) or amphipol (A-835, blue trace), monitored by UV absorbance. Experiment performed more than 3 times with similar results. Inset, calibration curve (n = 3 runs) demonstrating that Cx46/50 elutes at an apparent molecular weight of ~560 kDa, consistent with the size of a dodecameric protein complex (12 × ~38 kDa) and two micelles (2 × ~50 kDa). c, Electron micrograph of negatively stained Cx46/50 gap junctions reconstituted into amphipol. Scale bar, 100 nm. Inset, representative 2D class averages of negatively stained particles (selected from 25 classes). Scale bar, 20 nm. d–g, Chemical cross-linking and mass spectrometry. d, Representative MS/MS m/z spectrum, identifying inter-subunit cross-linking at the N-terminal Gly2 positions of Cx50 and Cx46. Identified peaks in the m/z spectrum and amino acid identities are indicated (Cx50 b-ions, green; Cx50 y-ions, yellow; Cx46 y-ions, blue). MS/MS data represent the consensus of 3 independent runs. e, Structural analysis of cross-linking results, showing inter-subunit distances between the symmetrically related N-terminal Gly2 positions within the connexin hemichannel, ranging from 9.1 Å (i to i ± 1), 15.8 Å (i to i ± 2) and 18.2 Å (i to i ± 3). The cross-linker spacer length is 11.4 Å, indicating a probable (i to i ± 1) arrangement of Cx50 and Cx46 within the same hemichannel, although other arrangements cannot be ruled out. f, Overview of identified inter-subunit cross-links between Cx50 and Cx46 assembled gap junctions. Residues in red indicate the site of primary amines involved in the cross-linking reaction using either DSS or BS3. All detected inter-subunit cross-links are between cytoplasmic domains. g, Schematic showing sites of inter-subunit cross-linking between Cx46 and Cx50 (black lines) and post-translational modifications identified during proteomics analysis (yellow, phosphorylation; purple, N-terminal acetylation). Met1 was determined to be removed in both Cx46 and Cx50 and the resulting N-terminal Gly2 position was identified in both acetylated and non-acetylated forms of Cx46 and Cx50, consistent with the specificity of the NatA acetylation complex24. The predicted CTD cleavage sites in Cx46 and Cx50 (orange), based on previous analysis of bovine Cx46/50 isolated from lens core tissue16, are also shown. Secondary structure and domain labels are indicated for the NTH, TM1–TM4, EC1, EC2, ICL and CTD.
Extended Data Fig. 2 Overview of cryo-EM image processing and 3D reconstruction.
a, A total of 1,104 micrographs were collected in an automated fashion using SerialEM65 on a 300-kV Titan Krios (dataset 1). Movie stacks were recorded using a K2 summit-direct electron detector operated in super-resolution mode and acquired with an effective pixel size of 0.665 Å. Movie stacks were corrected for drift and CTF using MotionCor266 and GCTF67, respectively. An initial dataset of 261,206 raw particles was obtained using unbiased autopicking procedures in DoG Picker68. A refined dataset of 53,791 good particles was obtained following several rounds of 2D classification and removal of ‘bad’ particles (or ice contamination) was done in Relion69. 3D classification was seeded using an initial model obtained by negative-stain electron microscopy, filtered to 60 Å. A majority of particles fell into a single 3D class (~62.5% of the good particles). These 30,128 particles were used for final 3D auto-refinement and post processing, yielding a final map at 3.4 Å resolution by gold-standard FSC (dataset 1). Dataset 2 was processed in a similar fashion from a total of 2,197 micrographs and 44,547 good particles, resulting in a final map at 3.5 Å resolution by gold-standard FSC. b, Summary of cryo-EM data collection, refinement and model validation statistics. Dataset 1 was used to obtain the 3.4 Å resolution reconstruction (map 1). Dataset 2 was used to obtain the 3.5 Å resolution reconstruction (map 2). Pre-processed and post-processed maps and associated masks from both datasets have been deposited in the Electron Microscopy Databank (EMD-9116). The original multi-frame micrographs have been deposited to EMPIAR (EMPIAR-10212). Coordinates for Cx50 and Cx46 atomic models have been deposited in the Protein Data Bank (6MHY and 6MHQ, respectively).
Extended Data Fig. 3 Global and local resolution analysis.
a, b, FSC analysis obtained from dataset 1 (a) and dataset 2 (b). Gold-standard FSC curves following auto-refinement (light grey), post-processing (grey), and masking (dark grey). The final masked maps display an overall resolution of ~3.4 Å (dataset 1) and ~3.5 Å (dataset 2), using a 0.143 cut-off. FSC curves comparing atomic models of Cx46 (orange) and Cx50 (blue) fit to the cryo-EM maps display correlation at 0.5 cut-off to a resolution of 3.4 Å (dataset 1) and 3.5 Å (dataset 2). c, d, Local resolution analysis using BlocRes70, obtained for the half-maps for dataset 1 (c) and 2 (d). e, f, Local resolution analysis comparing the experimental density map (dataset 1) to the calculated maps of Cx46 (e) and Cx50 (f). Local resolution ranges in c–f are indicated by colour (2.5–4.0 Å, blue–cyan; 4.0–5.0 Å, white; 5.0–6.5 Å, yellow–orange). Values obtained for local resolution of Cx46 and Cx50 models compared to the experimental density map are shown in Supplementary Tables 2, 3. Local resolution assessment comparing the density map to the two models indicates that the sites at which the two isoforms differ in sequence were generally less well-resolved, as compared to equivalently exposed residues at which Cx46 and Cx50 share conserved sequence.
Extended Data Fig. 4 Cx46 and Cx50 atomic models fit to the cryo-EM density maps.
Segmented cryo-EM map with atomic models for sheep Cx46 and Cx50 fit to the experimental densities derived from dataset 1 (3.4 Å, D6 symmetry), including regions for TM1–TM4, EC1 and EC2. The NTH domain is fit into the map from dataset 2 (3.5 Å, D6 symmetry), which was more well-defined in this region. Cx46 (top) and Cx50 (bottom) models are coloured according to their pairwise sequence homology, as being identical (grey, 80%), similar (blue, 8%) and different (orange, 12%). Windows show magnified views corresponding to boxed regions of the segmented maps, highlighting representative side-chain densities and fit to the atomic models. Regions of identical or similar amino acids are fit equally well by both models (for example, Cx46 L172 versus Cx50 I184, blue labels). Over regions in which the sequence of Cx46 and Cx50 differs, side-chain density is typically weaker (see also Extended Data Fig. 5). This is possibly due to the imposed averaging of two different side chains in these areas, or relative flexibility as many of these residues correspond to solvent-exposed side chains. In these areas of difference, and where electron microscopy density is present, both Cx46 and Cx50 models were typically fit equally well into the density map (for example, Cx46 T53 versus Cx50 V53 and Cx46 Q171 versus Cx50 R183, orange labels).
Extended Data Fig. 5 Analysis of different symmetry refinements on the ability to resolve differences between Cx46 and Cx50.
Eight sites of sequence differences involving bulky amino acids (labelled, and indicated by arrowhead) were selected as potential fiducial markers for resolving the two different isoforms following 3D refinement with various applied symmetries (C1, 4.1 Å resolution; C3, 3.9 Å resolution; C6, 3.7 Å resolution; D3, 3.7 Å resolution; D6, 3.4 Å resolution). For the applied symmetries, views are presented for each unique asymmetric subunit (boxed). Despite the modest resolution of the asymmetric (C1) reconstruction, side-chain density for bulky amino acids is typically observed at sites at which the two isoforms are conserved (asterisk). However, at the selected sites of sequence variation (arrow head) the side-chain densities are either not well-resolved, or there was no systematic variation that indicated an ability to distinguish the two isoforms. The most resolved features at these sites of variation were obtained with D6 symmetry, and typically corresponded to regions where these different amino acids share similar structure (such as Cβ positions). Although the cryo-EM density at these sites of variation were typically weak, the resolvable side-chain features throughout the rest of the map were generally enhanced when higher symmetry was applied during map refinement (indicated by asterisk), suggesting that regions of sequence similarity between Cx46 and Cx50 also share a high level of structural similarity.
Extended Data Fig. 6 Sequence and structure conservation of Cx46 and Cx50 heteromeric/heterotypic interfaces.
a, Multiple sequence alignment of mammalian Cx46 and Cx50 isoforms with residues contributing to heteromeric and heterotypic interfaces annotated83. Circle, heteromeric interface; square, heterotypic interface; filled, ≥70% buried; half-filled, 20–70% buried. Colouring corresponds to amino acid type (grey, hydrophobic; dark grey, aromatic; red, acidic; blue, basic; orange, hydrophilic; yellow, cysteine). Regions of sequence homology are indicated by the level of shading. Secondary structure and domain labels are indicated for the NTH domain, TM1–TM4, EC1 and EC2. Regions lacking defined structure and with poor sequence homology within the intracellular loop (ICL) and C-terminal domain (CTD) have been omitted for clarity. Sheep and human Cx46 and Cx50 orthologues contain ~95% sequence identity (~98% similarity) over the structured regions of the protein. Numbering corresponds to the amino acid sequence of sheep Cx44 and Cx49 used in the main text. b, Illustration of homomeric and heteromeric interface interactions involving the three sites lacking conservation between Cx44 and Cx49 at this interface (positions 39, 43 and 53). Despite these sequence differences, the interactions involving these residues are generally similar (hydrophobic, grey; hydrogen bonding, orange; ion-pairing, blue).
Extended Data Fig. 7 Analysis of protein backbone dynamics during molecular dynamics equilibration and production.
a, Cα r.m.s.d. analysis of equilibrium (0–30 ns) and production phases (30–80 ns) of the molecular dynamics simulations, calculated with respect to the experimental starting structure for non-acetylated models of Cx50 (red traces), Cx46 (blue traces) and Cx26 (with Met1 added; grey traces). Separate analysis for the NTH domains are shown in lighter shades. b, Same analysis as in a, for models with N-terminal acetylation added. The NTH domain of Cx26 (light grey traces) shows significantly higher r.m.s.d. values, for both non-acetylated and acetylated models. c, Plot of average Cα r.m.s.f. during the production phase of the molecular dynamics simulations for Cx50 (left, red traces), Cx46 (centre, blue traces) and Cx26 (right, grey traces). Data obtained for the N-terminal acetylated models are shown in lighter shades. Averages are determined for the 12 subunits composing the intercellular channel. Error bars represent 95% confidence intervals (n = 12 subunits). Secondary structure and domain labels are indicated for the NTH, TM1–4, EC1 and EC2, and ICL (not modelled). d, e, Average r.m.s.f. values of the acetylated models mapped to the experimental starting structures of Cx50 (left), Cx46 (centre) and Cx26 (right). Colours correspond to r.m.s.f. amplitudes: 0–1.0 Å (cyan); 1.0–2.0 Å (yellow–orange), >2.0 Å (red). In e, a single NTH domain is circled and the average r.m.s.f. values and 95% confidence intervals (Student t-test) calculated over the NTH domain of each isoform are displayed (n = 12 subunits). The NTH domain of Cx26 shows significantly higher r.m.s.f. values, for both acetylated and non-acetylated models (P < 0.0001).
Extended Data Fig. 8 Modulation of coulombic surface potential and K+/Cl− PMFs resulting from N-terminal acetylation and Cx46/50 heterotypic/heteromeric assembly.
a, Coulombic surface potential maps (top) and PMF (bottom) obtained for a set of non-acetylated and acetylated Cx50–Cx50ac and Cx46–Cx46ac models characterized by molecular dynamics simulation. Acetylated Cx50ac and Cx46ac monomers were used to construct a heterotypic channel and two different heteromeric channels (labelled I and II). Icons at the top of each structure show the relative configurations of Cx50 (black) and Cx46 (white). The resulting coulombic surface potentials are coloured as in Fig. 3a (negative, red; neutral, white; positive, blue). Only eight subunits are shown to portray both the channel pore and subunit interfaces. An asterisk (shown in a and b) indicates the site of N-terminal acetylation, which neutralizes the positively charged N terminus. PMFs obtained for K+ (blue traces) and Cl− ions (red traces) are displayed directly beneath each model displayed in a. Free-energy maxima are labelled and pore axis (z axis) is indicated. Traces indicate symmetrized values, with unsymmetrized values in lighter shading. In the case of the asymmetric Cx46/50 heterotypic model (middle left), PMFs represent the average from the first and last ~60 ns of simulation. b, Coulombic surface potential (top) and PMFs (bottom) obtained for non-acetylated Cx26 (left) and Cx26ac (right) are displayed as in a. c, Table of peak free-energy barriers for K+ (ΔGK+) and Cl− (ΔGCl−) and corresponding ΔΔG, reported as a proxy for charge selectivity. Asterisk indicates models with acetylated N terminus. d, e, Validation of methods used to construct the PMFs. d, Comparison of K+ PMFs obtained for Cx50ac and Cx46ac using population states (left) or transition rates (right) (see Methods). Both methods yielded similar PMF profiles. All other PMFs were constructed using the transition rates method. e, Comparison of Cl− PMFs obtained for Cx46ac using transition rates of Cl− ions that diffused into the pore (left) and those that were randomly seeded within the pore (right). All other Cl− PMFs were constructed using the distributed seeding approach to enhance sampling (see Methods).
Extended Data Fig. 9 Analysis of K+ trajectories and putative binding site observed during molecular dynamics simulation.
a–d, K+ ion trajectories obtained for Cx50 and Cx46 along the channel pore (z axis). a, b, Representative traces of mobile ions transiting and exiting or entering the channel pore in both acetylated (a) and non-acetylated (b) models of Cx46 and Cx50. c, d, K+ ions displaying long dwell times (~10–20 ns) localized at one or more putative binding sites within the channel pore (asterisk at z = ~14 Å) in both models of Cx46 and Cx50. In a–d, similar results were observed from 6 independent runs using non-acetylated models (1 × 80-ns and 2 × 10-ns runs for both Cx50 and Cx46) and 13 independent runs using the acetylated models (1 × 80-ns and 6 × 10-ns runs for Cx50; and 1 × 80-ns and 7 × 10-ns runs for Cx46). e, Representative snapshot showing an enlarged view of the putative K+ binding site identified for Cx50 and Cx46, corresponding to the region indicated by the asterisk in c and d. A single K+ ion is bound by a conserved set of amino acids (among Cx46/50 orthologues), coordinated by the side-chain carboxylate of Glu62 and backbone carbonyls of Gln49, Ser50 and Phe52 (identical in Cx46 and Cx50). Two transient water molecules observed coordinating the bond K+ ion are shown. Twelve binding sites are present within the dodecameric channel. Similar behaviour was observed from simulations using both non-acetylated and acetylated models (19 independent simulations). A functional role for this putative binding site is not yet clear, but may represent a physiologically relevant cation-binding site similar to the recently proposed Ca2+-binding site in Cx2611.
Extended Data Fig. 10 Dynamic hydrogen-bond network within the NTH domain observed by molecular dynamics simulation.
a–e, Analysis of hydrogen-bond interactions for non-acetylated models of Cx46 and Cx50 observed during molecular dynamics simulation. a, Inset, magnified view of D3 pairing with the positively charged N-terminal G2 position from a neighbouring subunit (intermolecular) and with the hydroxyl of S5 within the same subunit (intramolecular). The D3–G2 interactions are dynamically formed and broken during molecular dynamics simulation, whereas the intramolecular D3–S5 hydrogen bond is relatively stable (as shown in b–e). b, c, Population statistics of inter-atomic distances involving D3 (Cγ) and G (N) of the neighbouring chain (b) and Ser5 (Hγ) of the same chain (c), extracted from molecular dynamics simulation production runs of Cx46 (blue histogram) and Cx50 (red histogram). For D3 and G2, heavy atoms were chosen as proxies to monitor hydrogen-bonding interactions involving equivalent rotameric donor-acceptor configurations. The population centred at ~3.6 Å (b) and ~2.8 Å (c) are considered to be within hydrogen-bond distance. d, e, Trajectories extracted from molecular dynamics simulation of Cx46 (blue traces) and Cx50 (red traces) showing the dynamical behaviour of the D3–G2 intermolecular charge paring (d) and D3–S5 intramolecular hydrogen bonding (e). The dwell times showing hydrogen-bond pairing (~3.6 Å in d; and ~2.8 Å in e) are indicated with transparent grey shading. In the Cx26 crystal structure, the equivalent D2 site is modelled in hydrogen-bond distance to a neighbouring T5 site (Cx26 numbering)10, but this intermolecular interaction is rapidly broken during molecular dynamics simulations and does not appear to reform within the timescale of our molecular dynamics experiments, and instead forms a stable intramolecular interaction with T5, as previously described35 (data not shown). f–j, Analysis of hydrogen-bond interactions observed during molecular dynamics simulation for Cx46ac and Cx50ac modelled with the N-terminal G2 position acetylated. f, Inset, magnified view of acetylated G2ac position hydrogen bonded to the indole ring of W4 from the same subunit (intramolecular) and the same intramolecular D3–S5 hydrogen-bond interaction observed in the non-acetylated channel. g, h, Population statistics of inter-atomic distances involving W4 (Nε) and G2ac (acetyl carbonyl) (g) and D3 (Cγ) distance to Ser5 (Hγ) of the same chain (h), extracted from molecular dynamics simulation production runs of Cx46ac (blue histogram) and Cx50ac (red histogram). i, j, Trajectories extracted from molecular dynamics simulation of Cx46ac (blue traces) and Cx50ac (red traces) showing the dynamical behaviour of the W4–G2ac hydrogen-bond pairing (i) and D3–S5 intramolecular hydrogen bonding (j). The dwell times showing hydrogen-bond pairing (~2.4 Å in i, and ~2.5 Å in j) are indicated with transparent grey shading. For clarity, only the first 15 ns of the production period is shown (d, e, i, j). Similar results were observed from 6 independent runs using non-acetylated models (1 × 80-ns and 2 × 10-ns runs for both Cx50 and Cx46) and 13 independent runs using the acetylated models (1 × 80-ns and 6 × 10-ns runs for Cx50; and 1 × 80-ns and 7 × 10-ns runs for Cx46).
Supplementary information
Supplementary Information
This file contains: Supplementary Table 1: Summary of molecular dynamics simulation setup and conditions for Cx50, Cx46, Cx26 and Cx46/50 heteromeric/heterotypic models. Asterisk indicates models that included modification by n-terminal acetylation. Connexin-26 models were derived from PDB 2ZW310. Supplementary Table 2: Local resolution assessment of a calculated map based on the Cx46 atomic model versus the 3.4 Å resolution CryoEM density map. Values indicated by each residue correspond to the range of resolutions (Å) reported for amino acid sidechain densities using BlocRes70. Secondary structure and domain labels are indicated for the nterminal helix (NTH), transmembrane helices (TM1-4) and extracellular domains (EC1-2) intracellular loop (ICL) and C-terminal domain (CTD). Supplementary Table 3: Local resolution assessment of a calculated map based on the Cx50 atomic model versus the 3.4 Å resolution CryoEM density map. Values indicate by each residue correspond to the range of resolutions (Å) reported for amino acid sidechain densities using BlocRes70. Secondary structure and domain labels are indicated for the nterminal helix (NTH), transmembrane helices (TM1-4) and extracellular domains (EC1-2) intracellular loop (ICL) and C-terminal domain (CTD).
Rights and permissions
About this article
Cite this article
Myers, J.B., Haddad, B.G., O’Neill, S.E. et al. Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature 564, 372–377 (2018). https://doi.org/10.1038/s41586-018-0786-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0786-7
This article is cited by
-
Simulation of gap junction formation reveals critical role of Cys disulfide redox state in connexin hemichannel docking
Cell Communication and Signaling (2024)
-
Association of variants in GJA8 with familial acorea-microphthalmia-cataract syndrome
European Journal of Human Genetics (2024)
-
Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel
Nature Communications (2023)
-
Cryo-EM structure of human heptameric pannexin 2 channel
Nature Communications (2023)
-
Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.