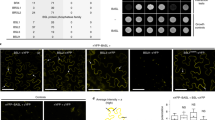
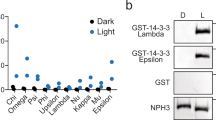
Abstract
Stomatal cell lineage is an archetypal example of asymmetric cell division (ACD), which is necessary for plant survival1,2,3,4. In Arabidopsis thaliana, the GLYCOGEN SYNTHASE KINASE3 (GSK3)/SHAGGY-like kinase BRASSINOSTEROID INSENSITIVE 2 (BIN2) phosphorylates both the mitogen-activated protein kinase (MAPK) signalling module5,6 and its downstream target, the transcription factor SPEECHLESS (SPCH)7, to promote and restrict ACDs, respectively, in the same stomatal lineage cell. However, the mechanisms that balance these mutually exclusive activities remain unclear. Here we identify the plant-specific protein POLAR as a stomatal lineage scaffold for a subset of GSK3-like kinases that confines them to the cytosol and subsequently transiently polarizes them within the cell, together with BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), before ACD. As a result, MAPK signalling is attenuated, enabling SPCH to drive ACD in the nucleus. Moreover, POLAR turnover requires phosphorylation on specific residues, mediated by GSK3. Our study reveals a mechanism by which the scaffolding protein POLAR ensures GSK3 substrate specificity, and could serve as a paradigm for understanding regulation of GSK3 in plants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets and accession codes supporting the findings of this study are available within the paper and its Supplementary Information. Source Data (gels and graphs) for Figs. 1–4 and Extended Data Figs. 1–10 are provided with the online version of the paper. There is no restriction on data availability.
References
Pillitteri, L. J., Guo, X. & Dong, J. Asymmetric cell division in plants: mechanisms of symmetry breaking and cell fate determination. Cell. Mol. Life Sci. 73, 4213–4229 (2016).
Pillitteri, L. J. & Dong, J. Stomatal development in Arabidopsis. Arabidopsis Book 11, e0162 (2013).
MacAlister, C. A., Ohashi-Ito, K. & Bergmann, D. C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 (2007).
Pillitteri, L. J., Peterson, K. M., Horst, R. J. & Torii, K. U. Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23, 3260–3275 (2011).
Kim, T.-W., Michniewicz, M., Bergmann, D. C. & Wang, Z.-Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422 (2012).
Khan, M. et al. Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J. Biol. Chem. 288, 7519–7527 (2013).
Gudesblat, G. E. et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548–554 (2012).
Lampard, G. R., MacAlister, C. A. & Bergmann, D. C. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116 (2008).
Lampard, G. R., Lukowitz, W., Ellis, B. E. & Bergmann, D. C. Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21, 3506–3517 (2009).
Lin, G. et al. A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 31, 927–938 (2017).
Dong, J., MacAlister, C. A. & Bergmann, D. C. BASL controls asymmetric cell division in Arabidopsis. Cell 137, 1320–1330 (2009).
Zhang, Y., Wang, P., Shao, W., Zhu, J.-K. & Dong, J. The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev. Cell 33, 136–149 (2015).
Zhang, Y., Guo, X. & Dong, J. Phosphorylation of the polarity protein BASL differentiates asymmetric cell fate through MAPKs and SPCH. Curr. Biol. 26, 2957–2965 (2016).
Youn, J.-H. & Kim, T.-W. Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8, 552–565 (2015).
Kim, T.-W. et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260 (2009).
Yan, Z., Zhao, J., Peng, P., Chihara, R. K. & Li, J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150, 710–721 (2009).
Rozhon, W., Mayerhofer, J., Petutschnig, E., Fujioka, S. & Jonak, C. ASKθ, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 62, 215–223 (2010).
De Rybel, B. et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16, 594–604 (2009).
Vert, G. & Chory, J. Downstream nuclear events in brassinosteroid signalling. Nature 441, 96–100 (2006).
Blomme, J. et al. The mitochondrial DNA-associated protein SWIB5 influences mtDNA architecture and homologous recombination. Plant Cell 29, 1137–1156 (2017).
Nadeau, J. A. & Sack, F. D. Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700 (2002).
Wakefield, J. G., Stephens, D. J. & Tavaré, J. M. A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment. J. Cell Sci. 116, 637–646 (2003).
Adrian, J. et al. Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev. Cell 33, 107–118 (2015).
Yang, K.-Z. et al. Phosphorylation of serine 186 of bHLH transcription factor SPEECHLESS promotes stomatal development in Arabidopsis. Mol. Plant 8, 783–795 (2015).
Kondo, Y. et al. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF–TDR signalling. Nat. Commun. 5, 3504 (2014).
Hecker, A. et al. Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiol. 168, 776–787 (2015).
Wang, X. & Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122 (2006).
Fauser, F., Schiml, S. & Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359 (2014).
Lampropoulos, A. et al. GreenGate—a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8, e83043 (2013).
Karimi, M., De Meyer, B. & Hilson, P. Modular cloning in plant cells. Trends Plant Sci. 10, 103–105 (2005).
Lei, Y. et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496 (2014).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Edwards, K., Johnstone, C. & Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349 (1991).
Peterson, K. M. & Torii, K. U. Long-term, high-resolution confocal time lapse imaging of Arabidopsis cotyledon epidermis during germination. J. Vis. Exp. 70, e4426 (2012).
Delgado, D., Alonso-Blanco, C., Fenoll, C. & Mena, M. Natural variation in stomatal abundance of Arabidopsis thaliana includes cryptic diversity for different developmental processes. Ann. Bot. 107, 1247–1258 (2011).
Andriankaja, M. et al. Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22, 64–78 (2012).
Boruc, J. et al. Functional modules in the Arabidopsis core cell cycle binary protein–protein interaction network. Plant Cell 22, 1264–1280 (2010).
Wendrich, J. R., Boeren, S., Möller, B. K., Weijers, D. & De Rybel, B. In vivo identification of plant protein complexes using IP–MS/MS. Methods Mol. Biol. 1497, 147–158 (2017).
Van Leene, J. et al. An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10, 169–187 (2015).
Nelissen, H. et al. Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27, 1605–1619 (2015).
Acknowledgements
We thank D. Bergmann, K. Torii, C. Grefen, Y. Kondo and H. Fukuda for materials; M. Pfeiffer, B. Pavie, J. Winkler, D. Van Damme, E. Mylle, S. Dhondt and A. Baekelandt for microscopy; M. De Cock for editing; V. Storme for statistics; B. De Rybel, J. Friml and N. Raikhel for critical reading. This work was supported by the Research Foundation—Flanders (G008416N) (E.R.), the China Scholarship Council (C.Z., K.W.), the Belgian Science Policy (K.W., M.T., M.K.Z., G.E.G.), the Agency for Innovation by Science and Technology (IWT) (M.K.Z.) and the Spanish Government Research Grant AGL2015-65053-R (M.M., C.F.).
Reviewer information
Nature thanks S. Casson and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.H., C.Z., M.T., K.W., A.d.M.S., D.V.S., M.J.U., M.K.Z., G.E.G., I.V., D.E., S.B., M.K. and C.B. performed experiments. T.J., C.F., M.M., S.d.V., G.D.J., A.H. and E.R. designed experiments. A.H. and E.R. wrote the manuscript and all authors revised it.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 In vivo complex formation and self-polarization of BIN2, ATSK12, POLAR and BASL.
a, Transient expression in tobacco leaf epidermis of BIN2–GFP, ATSK12–GFP, POLAR–mCherry and mCherry–BASL. b, Transient co-expression of BIN2–GFP with POLAR–mCherry without polarization of either of the proteins. c, Transient co-expression of BIN2–GFP with mCherry–BASL inducing some BIN2 polarization. d, Transient co-expression of POLAR–GFP with mCherry–BASL strongly polarizing POLAR. e, Transient co-expression of BIN2–GFP with POLAR–mCherry and TagBFP2–BASL strongly polarizing BIN2 and POLAR. f, Transient co-expression of ATSK12–GFP with POLAR–mCherry and TagBFP2–BASL strongly polarizing ATSK12 and POLAR. g, Quantification of BIN2, POLAR and BASL polarity. n, number of cells from three biologically independent leaves. The polarity was determined by the ratio between the average intensities at the high (H) and low (L) polarity regions. For quantification, all proteins were co-expressed with the plasma membrane marker PM-TagBFP2. White arrowheads in a–f indicate polarity in the lobes. For a–f, experiments were repeated independently three times. Box plots show the first and third quartiles, split by the median (line) and mean (cross). Statistical analysis was done with a one-way ANOVA and Tukey’s post hoc test compared to the respective single infiltration. Scale bars, 20 µm.
Extended Data Fig. 2 Nuclear exclusion of BIN2 by POLAR and of BASL by BIN2.
a, Transient expression in tobacco leaf epidermis of BIN2–GFP, POLAR–mCherry, mCherry–BASL, and the kinase-dead version of BIN2 (BIN2(K69R)–GFP). b, Transient co-expression of BIN2–GFP or BIN2(K69R)–GFP with POLAR–mCherry excluding BIN2 from the nucleus. c, Transient co-expression of BIN2–GFP with mCherry–BASL excluding BASL from the nucleus, whereas the transient co-expression of BIN2(K69R)–GFP with mCherry–BASL did not. The nucleus is indicated with arrowheads (a–c). d, Quantification of the nuclear exclusion of BIN2 and BASL. n, number of cells from three biologically independent leaves. Images were quantified with the nuclear (N)-TagBFP2 as a marker for the nucleus area and by means of a script to measure the average intensity in the nucleus compared to the remainder of the cell (see Methods). For a–c, experiments were repeated independently three times. Box plots show the first and third quartiles, split by the median (line) and mean (cross). Statistical analysis was performed with a one-way ANOVA and Tukey’s post hoc test compared to BIN2–GFP or BIN2(K69R) or mCherry–BASL single infiltration, respectively. Scale bars, 20 µm.
Extended Data Fig. 3 Nuclear exclusion of other members of the Arabidopsis GSK3/SHAGGY family by POLAR.
a, b, Transient co-expression in tobacco leaf epidermis of POLAR–mCherry with the group-I GSK3/SHAGGY-like kinases ATSK11(ASKα)–GFP, ATSK12(ASKγ)–GFP, and ATSK13(ASKε)–GFP (a) and with the group-II GSK3/SHAGGY-like kinases BIN2(ASKη)–GFP, BIL2(ASKι)–GFP, and (BIL1)ASKζ–GFP (b) leading to their nuclear exclusion. c, Transient co-expression of POLAR–mCherry with the group-III GSK3/SHAGGY-like kinases ATSK32(ASKθ)–GFP and ATSK31(ASKβ)–GFP. ATSK31(ASKβ)–GFP was excluded from the nucleus, whereas ATSK32(ASKθ)–GFP was not; POLAR–mCherry was included into the nucleus with ATSK32(ASKθ)–GFP. d, Transient co-expression of POLAR–mCherry with the group-IV GSK3/SHAGGY-like kinases ATSK41(ASKκ)–GFP and ATSK42(ASKδ)–GFP. When expressed alone, ATSK41(ASKκ)–GFP and ATSK42(ASKδ)–GFP did not localize to the nucleus, and coinfiltration with POLAR–mCherry did not affect their localization. White arrowheads in a–d indicate the nucleus. For a-d, experiments were repeated independently three times. Scale bars, 20 µm.
Extended Data Fig. 4 Characterization of Arabidopsis Col-0 and atsk12 mutant endogenously expressing BIN2–GFP and ATSK12–GFP, respectively.
a, Wild-type (Col-0) and BIN2:BIN2-GFP;Col-0 plants (lines 21 and 26), grown for 4 weeks in soil with corresponding BIN2 gene expression in seedlings measured by qRT–PCR. n = 3 biologically independent experiments. Scale bars, 2.5 cm b, Schematic representation of the tDNA insertions in the atsk12 mutant (GABI-410D09). The insertion site is annotated with the flanking sequence and corresponding insertion site or inserted nucleotide in green. Primers used are depicted in the scheme and listed in Supplementary Table 2. Wild-type (Col-0) and ATSK12:ATSK12-GFP;atsk12 complemented plants (lines 2, 3 and 14) grown for 2 weeks in soil with the corresponding ATSK12 gene expression in seedlings measured by qRT–PCR. n = 3 biologically independent experiments. All qRT–PCR were carried out with seedlings at 5 d.p.g. Transcript levels were normalized to the ACTIN2 gene expression. c, In vitro phosphorylation of YDA and SPCH by ATSK12. All experiments were repeated independently three times. Box plots show the first and third quartiles, split by the median (line) and mean (cross). For blot source data, see Supplementary Fig. 1.
Extended Data Fig. 5 Localization of BIN2, ATSK12, BASL and POLAR in Arabidopsis.
a, Localization of BIN2 in the abaxial epidermis of Arabidopsis seedlings at 1, 2 or 5 d.p.g. BIN2 was not detectable at 1 d.p.g., but was visible at 1.5–2 d.p.g. BIN2 localized to the plasma membrane (PM), cytoplasm and nucleus. BIN2 was more abundant and polarized at the cell cortex before every ACD. b, Localization of ATSK12 in the abaxial epidermis of Arabidopsis seedlings at 1, 2 or 5 d.p.g. ATSK12 was detected at all developmental stages and localized to the plasma membrane and cytoplasm, but unlike BIN2, it had no nuclear signal. ATSK12 was also more abundant and polarized at the cell cortex before every ACD. Insets in a and b depict BIN2 and ATSK12 localization during cytokinesis. During mitosis, both proteins relocalized to two distinct foci resembling the spindle localization as observed for mammalian GSK3. c, Localization of BASL in the abaxial epidermis of Arabidopsis seedlings at 1 d.p.g. d, Localization of POLAR in the abaxial epidermis of Arabidopsis seedlings at 1 d.p.g. e, BIN2 localization in POLAR loss- and gain-of-function mutants. BIN2 abundance in stomatal lineage cells undergoing ACD depends on POLAR. Loss of POLAR and PL1 markedly reduced, whereas POLAR overexpression greatly enhanced, BIN2 enrichment before the ACD. Insets represent zoomed in regions. f, Quantification of the abundance of BIN2–GFP in the plasma membrane of cells undergoing ACD. n, number of cells from three biologically independent cotyledons. The intensity of BIN2–GFP was measured over time in the plasma membrane in stomatal lineage cells undergoing ACD. The plasma membrane signal of meristemoid cells was divided by the plasma membrane signal of neighbouring differentiated pavement cells to quantify the relative protein abundance. g, Nuclear localization of BIN2–GFP in Col-0 and the double tDNA polar-2 pl1-1 mutant in the abaxial epidermis at 2 d.p.g. Meristemoids of the double polar-2 pl1-1 mutant showed the increased nuclear localization of BIN2–GFP . The pictures below each main image represent zoomed in regions. White arrowheads indicate the nucleus in meristemoids. h, Quantification of the nuclear signal for BIN2 in g. n, number of cells from three biologically independent cotyledons. All experiments were repeated independently three times. Box plots show the first and third quartiles, split by the median (line) and mean (cross). One-way ANOVA with Tukey’s post hoc test was used as indicated by brackets. Scale bars, 20 µm.
Extended Data Fig. 6 POLAR stabilized group I and group II ATSKs in the stomatal lineage.
a, Confocal images of abaxial epidermis of cotyledons of seedlings at 3 d.p.g. expressing group-I ATSK11:ATSK11-GFP;Col-0 (AT5G26751), ATSK12:ATSK12-GFP;Col-0 (AT3G05840) or ATSK13:ATSK13-GFP (AT5G14640) and group-II BIN2:BIN2-GFP;Col-0, BIL2:BIL2-GFP;Col-0 (AT1G06390) and BIL1:BIL1-GFP;Col-0 (AT2G30980). ATSK11–GFP, ATSK12–GFP, ATSK13–GFP, BIN2–GFP, BIL2–GFP and BIL1–GFP proteins showed stabilization in the stomatal lineage and polarization during ACD, with group-I members displaying a more frequent polarization. b, Co-expression of all BIN2 homologues with POLAR–mCherry leading to their stabilization and apparent polarization. White arrowheads in a and b indicate stabilization and/or polarization and correspond to insets in every picture. c, Co-expression of BIN2 with mCherry–BASL did not lead to BIN2 stabilization and polarization. All experiments were repeated independently three times. Scale bars, 20 µm.
Extended Data Fig. 7 Identification and characterization of POLAR, POLAR-like1 (PL1) and PL2 loss-of-function mutants.
a–c, Schematic representation of the tDNA insertions in the polar-2 (SALK_146639) (a), pl1-1 (SALK_121087) (b) and pl2-1 (SALK_123929) (c) mutants. d, Schematic representation of the POLAR and PL1 CRISPR mutants polar-3 and pl1-2, respectively, and the double CRISPR mutant polar-4 pl1-2. Guide sequences of the selected target sites of the wild-type and of the CRISPR-edited polar are presented under the scheme. In the boxes, the protospacer-adjacent motif sequences are shown in red and inserted or deleted nucleotides in green. The resulting truncated amino acid sequence of the protein is shown below the scheme. Amino acids in green correspond to the wild-type POLAR protein and those in red to random amino acids produced by the frame shift after the nucleotide insertion/deletion. e, Wild-type (Col-0), POLAR tDNA (polar-2) and POLAR CRISPR (polar-3) mutants and POLAR-like1 (PL1) tDNA (pl1-1) and PL1 CRISPR (pl1-2) mutants, as well as the corresponding double tDNA (polar-2 pl1-1) and CRISPR (polar-4 pl1-2) mutants grown for 4 weeks in soil. Scale bars, 2.5 cm. f, POLAR, PL1 and PL2 gene expression measured by qRT–PCR in seedlings of polar-2, pl1-1 and pl2-1 plants. n = 3 biologically independent experiments. Primers used are depicted in the scheme and listed in Supplementary Table 2. For e and f, experiments were repeated independently three times. Box plots show the first and third quartiles, split by the median (line) and mean (cross).
Extended Data Fig. 8 BIN2 and POLAR overexpression in the stomatal lineage of Arabidopsis.
a, TMM:BIN2-GSgreen;bin2-3 (lines 17.6 and 16.3), TMM:BIN2-GSgreen;Col-0 (lines 4.2 and 6.2), POLAR:POLAR-GFP;Col-0 (lines 14.5 and 16.3) and POLAR:POLAR-mCherry (lines 6 and 5) introduced into TMM:BIN2-GSgreen;Col-0 (line 6.2). Scale bar, 2.5 cm. b, BIN2 and POLAR gene expression in the genotypes shown in a, measured by qRT–PCR. n = 3 biologically independent experiments. c, SPCH–mCherry marking the stomatal lineage cell fate of the small dividing cells in pTMM:BIN2-GSgreen;Col-0 (line 6.2) plants. d, Localization of POLAR–GFP in plants overexpressing POLAR-GFP;Col-0 (line 14.5) presenting excessive divisions in the stomatal lineages. All experiments were repeated independently three times. Box plots show the first and third quartiles, split by the median (line) and mean (cross). Scale bars in c and d, 20 µm.
Extended Data Fig. 9 Stomatal phenotypes in abaxial epidermis of cotyledons at 21 d.p.g.
a, Wild types (Col-0 or Wassilewskija (Ws)), polar-2, pl1-1, polar-2 pl1-1, polar-3 (CRISPR), pl1-2 (CRISPR), polar-4 pl1-2 (CRISPR), POLAR:POLAR-GFP;Col-0 (line 14.5), bin2-3, atsk12, bin2-3 atsk12, bin2-3 bil1 bil2 atsk13-RNAi, TMM:BIN2-GSgreen;bin2-3 (line 17.6), TMM:BIN2-GSgreen;Col-0 (line 6.2), TMM:BIN2-GSgreen;POLAR:POLAR-mCherry (line 6), and basl-2 seedlings. Scale bar, 100 µm. b, Pavement cell size distribution of genotypes described in a with significant phenotypes. n, number of cells from three biologically independent cotyledons. c, Quantification of the pavement cell density, stomatal density, total cell density, the stomatal index, and the stomatal clustering index in the genotypes described in a. n, number of biologically independent cotyledons. Box plots show the first and third quartiles, split by the median (line) and mean (cross). Statistical analysis was done with a one-way ANOVA and Tukey’s post hoc test. All values were compared to Col-0 unless indicated with brackets. The bin2-3 bil1 bil2 atsk13-RNAi quadruple mutant was compared to Ws. For a–c, experiments were repeated independently three times.
Extended Data Fig. 10 Phosphorylation of BASL by BIN2 and the working model for BIN2 and POLAR functions during ACD.
a, In vitro phosphorylation of BASL by BIN2. Alanine substitutions in five BIN2 phosphorylated residues (BASL(5A)) reduced the BASL phosphorylation. The S72A substitution in the truncated BASL (BASL(1–84/S72A)) abolished phosphorylation. b, BASL protein. The blue dots, red asterisks, and green dot mark the identified in vitro BIN2 phosphorylation sites (this study), previously described MAPK phosphorylation sites, and the predicted S72 phosphorylation site, respectively. The nuclear localization signal (NLS) is marked in blue. The putative GSK3 phosphorylation motifs are marked in yellow. c, Confocal images of abaxial epidermis of cotyledons at 2.5 d.p.g. of BASL:GFP-BASL;basl-2 or SPCH:SPCH-GFP;spch-3 plants grown in the presence of 50 μM bikinin (BIK) or mock (DMSO). Scale bars, 20 μm. d, Wild-type (Col-0) seedlings at 2.5 d.p.g. grown in the presence of 50 μm BIK or mock (DMSO). Western blot analysis of MPK3 and MPK6 activation upon treatment with BIK. Active MPK3 and MPK6 were detected by a phospho-p44/42 MAPK antibody. For blot source data, see Supplementary Fig. 1. e, Gene expression of SPCH, POLAR and BASL as in d, measured by qRT–PCR. n = 3 biologically independent experiments. Transcript levels were normalized to the UBIQUITIN10 gene. f, Model for the function of BIN2 and POLAR during ACDs. In the MMC or young meristemoid (M), POLAR is highly expressed, nonpolar, whereas the BIN2 expression is absent or undetectable. In mature MMCs or meristemoids, BIN2 is expressed, is upregulated and initiates BASL polarization redundantly with the MAPK module. The BIN2–POLAR–BASL complex migrates to the cell cortex and polarizes before the ACD, thereby relieving the BIN2 inhibition on SPCH in the nucleus and attenuating the MAPK signalling in the polar region. After the ACD, the smaller daughter cell (meristemoid) accumulates SPCH to sustain POLAR and BASL transcription, allowing further amplifying ACDs. When the POLAR expression remains in the larger daughter cell (SLGC), the BIN2–POLAR–BASL polarity module is reoriented at the opposite side, allowing the occurrence of spacing divisions. When the POLAR expression in the SLGC is low, BIN2 is more abundant in the nucleus, relieving its inhibitory function on the MAPK signalling, leading to SPCH degradation and pavement cell differentiation. Meristemoids destined for a guard mother cell fate lose the POLAR expression, while maintaining the BIN2 expression. For a–e, experiments were repeated independently three times. Box plots show the first and third quartiles, split by the median (line) and mean (cross).
Supplementary information
Supplementary Figures
This file contains Supplementary Figure 1: Source data for images for gels and blots.
Supplementary Table
This file contains Supplementary Table 1: Mass spectrometry (MS) analysis.
Supplementary Table
This file contains Supplementary Table 2: Primer list used in this study.
Video 1: Confocal time-lapse imaging of BASL–GFP
Confocal time-lapse imaging of BASL:GFP-BASL;basl-2 in the abaxial epidermis of cotyledons of Arabidopsis seedlings at 2 days post germination. In meristemoids and stomatal lineage ground cells (SLGC) that will undergo asymmetric cell division (ACD), BASL first localizes to the nucleus and then it polarizes at the cell cortex preceding the ACD. The polarity is only inherited in the SLGC and is further enhanced between 30 to 60 min after ACD. Experiments were repeated independently three times.
Video 2: Confocal time-lapse imaging of POLAR–GFP
Confocal time-lapse imaging of POLAR:POLAR-GFP;polar-2 in the abaxial epidermis of cotyledons of Arabidopsis seedlings at 2 days post germination. In meristemoids and stomatal lineage ground cells (SLGC) that will undergo asymmetric cell division (ACD), POLAR first localizes evenly to the plasma membrane, then is upregulated and polarizes before ACD. The polarity is inherited in the SLGC but is the strongest prior to ACD, unlike BASL. Experiments were repeated independently three times.
Video 3: Confocal time-lapse imaging of BIN2–GFP
Confocal time-lapse imaging of pBIN2-BIN2-GFP/BIN2:BIN2-GFP;Col-0 in the abaxial epidermis of cotyledons of Arabidopsis seedlings at 2 days post germination. In meristemoids and stomatal lineage ground cells (SLGC) that will undergo asymmetric cell division (ACD), BIN2 first localizes evenly to the plasma membrane, then is upregulated and polarizes before ACD. The polarity is only weakly inherited in the SLGC and just like for POLAR, the polarity shows a maximum before ACD. Experiments were repeated independently three times.
Video 4: Confocal time-lapse imaging of ATSK12–GFP
Confocal time-lapse imaging of ATSK12:ATSK12-GFP;atsk12 in the abaxial epidermis of cotyledons of Arabidopsis seedlings at 2 days post germination. In meristemoids and stomatal lineage ground cells (SLGC) that will undergo asymmetric cell division (ACD), AtSK12 first localizes evenly to the plasma membrane, then is upregulated and polarizes before ACD. The polarity is only weakly inherited in the SLGC and just like for POLAR, the polarity shows a maximum before ACD. Experiments were repeated independently three times.
Video 5: Confocal time-lapse imaging of BIN2–GFP when POLAR is overexpressed
Confocal time-lapse imaging of BIN2:BIN2-GFP;POLAR:POLAR-mCherry;Col-0 in the abaxial epidermis cotyledons of Arabidopsis seedlings at 2 days post germination. In meristemoids and stomatal lineage ground cells (SLGC) that will undergo asymmetric cell division (ACD), BIN2 localizes evenly to the plasma membrane, then is strongly upregulated and polarized before ACD. The polarity and enrichment of BIN2 prior to ACD is amplified by the overexpression of POLAR. Experiments were repeated independently three times.
Video 6: Confocal time-lapse imaging of BIN2–GFP in a polar-2 pl1-1 double mutant
Confocal time-lapse imaging of BIN2:BIN2-GFP;polar-2 pl1-1 in the abaxial epidermis cotyledons of Arabidopsis seedlings at 2 days post germination. In meristemoids and stomatal lineage ground cells (SLGC) that will undergo asymmetric cell division (ACD), BIN2 localizes evenly to the plasma membrane but fails to be enriched and polarized before ACD. The polarity and enrichment of BIN2 is dependent on POLAR. Experiments were repeated independently three times.
Source data
Rights and permissions
About this article
Cite this article
Houbaert, A., Zhang, C., Tiwari, M. et al. POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563, 574–578 (2018). https://doi.org/10.1038/s41586-018-0714-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0714-x
Keywords
This article is cited by
-
Adaptor protein complex interaction map in Arabidopsis identifies P34 as a common stability regulator
Nature Plants (2023)
-
The heat is on: a simple method to increase genome editing efficiency in plants
BMC Plant Biology (2022)
-
Dichotomy of the BSL phosphatase signaling spatially regulates MAPK components in stomatal fate determination
Nature Communications (2022)
-
Dynamic chromatin accessibility deploys heterotypic cis/trans-acting factors driving stomatal cell-fate commitment
Nature Plants (2022)
-
Connected function of PRAF/RLD and GNOM in membrane trafficking controls intrinsic cell polarity in plants
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.