Abstract
Pathogenic bacteria such as Escherichia coli assemble surface structures termed pili, or fimbriae, to mediate binding to host-cell receptors1. Type 1 pili are assembled via the conserved chaperone–usher pathway2,3,4,5. The outer-membrane usher FimD recruits pilus subunits bound by the chaperone FimC via the periplasmic N-terminal domain of the usher. Subunit translocation through the β-barrel channel of the usher occurs at the two C-terminal domains (which we label CTD1 and CTD2) of this protein. How the chaperone–subunit complex bound to the N-terminal domain is handed over to the C-terminal domains, as well as the timing of subunit polymerization into the growing pilus, have previously been unclear. Here we use cryo-electron microscopy to capture a pilus assembly intermediate (FimD–FimC–FimF–FimG–FimH) in a conformation in which FimD is in the process of handing over the chaperone-bound end of the growing pilus to the C-terminal domains. In this structure, FimF has already polymerized with FimG, and the N-terminal domain of FimD swings over to bind CTD2; the N-terminal domain maintains contact with FimC–FimF, while at the same time permitting access to the C-terminal domains. FimD has an intrinsically disordered N-terminal tail that precedes the N-terminal domain. This N-terminal tail folds into a helical motif upon recruiting the FimC-subunit complex, but reorganizes into a loop to bind CTD2 during handover. Because both the N-terminal and C-terminal domains of FimD are bound to the end of the growing pilus, the structure further suggests a mechanism for stabilizing the assembly intermediate to prevent the pilus fibre diffusing away during the incorporation of thousands of subunits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The cryo-EM 3D maps of the FimD–tip (FimDCFGH) complex have been deposited at the Electron Microscopy Data Bank database with accession codes EMD-8953 (conformer 1) and EMD-8954 (conformer 2), and their corresponding atomic models were deposited at the PDB with accession codes 6E14 (conformer 1) and 6E15 (conformer 2), respectively.
References
Flores-Mireles, A. L., Walker, J. N., Caparon, M. & Hultgren, S. J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284 (2015).
Thanassi, D. G., Saulino, E. T. & Hultgren, S. J. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1, 223–231 (1998).
Geibel, S. & Waksman, G. The molecular dissection of the chaperone–usher pathway. Biochim. Biophys. Acta 1843, 1559–1567 (2014).
Zav’yalov, V., Zavialov, A., Zav’yalova, G. & Korpela, T. Adhesive organelles of Gram-negative pathogens assembled with the classical chaperone/usher machinery: structure and function from a clinical standpoint. FEMS Microbiol. Rev. 34, 317–378 (2010).
Nuccio, S. P. & Bäumler, A. J. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 (2007).
Mulvey, M. A. et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282, 1494–1497 (1998).
Sauer, F. G. et al. Chaperone-assisted pilus assembly and bacterial attachment. Curr. Opin. Struct. Biol. 10, 548–556 (2000).
Choudhury, D. et al. X-ray structure of the FimC–FimH chaperone–adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066 (1999).
Sauer, F. G. et al. Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061 (1999).
Zavialov, A. V. et al. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113, 587–596 (2003).
Sauer, F. G., Pinkner, J. S., Waksman, G. & Hultgren, S. J. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111, 543–551 (2002).
Nishiyama, M., Ishikawa, T., Rechsteiner, H. & Glockshuber, R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379 (2008).
Phan, G. et al. Crystal structure of the FimD usher bound to its cognate FimC–FimH substrate. Nature 474, 49–53 (2011).
Nishiyama, M. et al. Structural basis of chaperone–subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 24, 2075–2086 (2005).
Ng, T. W., Akman, L., Osisami, M. & Thanassi, D. G. The usher N terminus is the initial targeting site for chaperone–subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186, 5321–5331 (2004).
Saulino, E. T., Thanassi, D. G., Pinkner, J. S. & Hultgren, S. J. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 17, 2177–2185 (1998).
Volkan, E. et al. Domain activities of PapC usher reveal the mechanism of action of an Escherichia coli molecular machine. Proc. Natl Acad. Sci. USA 109, 9563–9568 (2012).
Nishiyama, M. & Glockshuber, R. The outer membrane usher guarantees the formation of functional pili by selectively catalyzing donor-strand exchange between subunits that are adjacent in the mature pilus. J. Mol. Biol. 396, 1–8 (2010).
Rose, R. J. et al. Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc. Natl Acad. Sci. USA 105, 12873–12878 (2008).
Geibel, S., Procko, E., Hultgren, S. J., Baker, D. & Waksman, G. Structural and energetic basis of folded-protein transport by the FimD usher. Nature 496, 243–246 (2013).
Allen, W. J., Phan, G., Hultgren, S. J. & Waksman, G. Dissection of pilus tip assembly by the FimD usher monomer. J. Mol. Biol. 425, 958–967 (2013).
Eidam, O., Dworkowski, F. S., Glockshuber, R., Grütter, M. G. & Capitani, G. Crystal structure of the ternary FimC–FimFt–FimDN complex indicates conserved pilus chaperone–subunit complex recognition by the usher FimD. FEBS Lett. 582, 651–655 (2008).
Crespo, M. D. et al. Quality control of disulfide bond formation in pilus subunits by the chaperone FimC. Nat. Chem. Biol. 8, 707–713 (2012).
Werneburg, G. T. et al. The pilus usher controls protein interactions via domain masking and is functional as an oligomer. Nat. Struct. Mol. Biol. 22, 540–546 (2015).
Volkan, E. et al. Molecular basis of usher pore gating in Escherichia coli pilus biogenesis. Proc. Natl Acad. Sci. USA 110, 20741–20746 (2013).
Remaut, H. et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652 (2008).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Henderson, N. S., Ng, T. W., Talukder, I. & Thanassi, D. G. Function of the usher N-terminus in catalysing pilus assembly. Mol. Microbiol. 79, 954–967 (2011).
Acknowledgements
Cryo-EM data were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite at the Van Andel Research Institute. We thank X. Meng for help with data collection. This study was supported by the US National Institutes of Health R01 grants GM062987 (to D.G.T. and H.L.) and GM111742 (to H.L.) and Van Andel Research Institute (to H.L.).
Reviewer information
Nature thanks A. Dessen, J. Rubinstein and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
D.G.T. and H.L. conceived and designed experiments. M.D., N.H., S.S. and G.T.W. carried out biochemical and molecular biology experiments. M.D., Z.Y. and G.Z. performed cryo-EM experiments. M.D., Z.Y. and H.Y. performed image processing and atomic modelling. M.D., Z.Y., H.Y, D.G.T. and H.L. analysed the data. M.D., Z.Y., H.Y., D.G.T. and H.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Pilus assembly occurs via donor-strand complementation and donor-strand exchange.
This sketch is based on the crystal structure of FimD–FimC–FimF–FimG–FimH (PDB ID 4J3O). Donor-strand complementation (DSC): pilus subunits (FimF, in this case) have an immunoglobulin-like structure in which the C-terminal G strand is missing. In the periplasm, the chaperone FimC donates its G1 strand to complete the subunit fold, but in a non-canonical parallel orientation with the subunit F strand. Donor-strand exchange (DSE): the donor strand of the FimC chaperone in the previous subunit (FimG, in this case) is replaced by the N-terminal extension of the incoming subunit (FimF), completing the FimG-subunit immunoglobulin fold in a canonical, anti-parallel orientation.
Extended Data Fig. 2 Cryo-EM of FimD–tip complex.
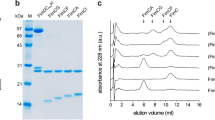
a, The gel filtration profile of FimD–tip complex from a Superdex 200 10/300GL column. b, Coomassie blue SDS–PAGE gel of the peak fraction, which shows the presence of all subunits of the purified FimD–tip complex. Similar sample preparations by gel filtration and SDS–PAGE examination were carried out more than three times. c, A raw cryo-EM micrograph of the purified FimD–tip complexes embedded in vitreous ice. A total of 12,000 such micrographs was recorded. d, Selected 2D class averages showing the presence of many different views and well-resolved structural features. Over 750,000 raw particles contributed to final 2D class averages. e, Three-dimensional classification scheme. Over 1 million raw particles were selected from drift-corrected electron micrographs. Two- and three-dimensional classification resulted in two 3D maps that were of the expected shape, and the structure appeared complete; the other three maps were either partial structures or distorted. Refinement with about 250,370 particles led to the 4.0-Å resolution 3D map of conformer 1, and refinement with about 166,913 particles led to the 5.1-Å resolution 3D map of conformer 2.
Extended Data Fig. 3 Resolution estimation and selected regions of the 3D electron microscopy maps.
a, Local-resolution estimation and the Gold-standard Fourier shell correlation estimation at the 0.143 correlation threshold of the FimD–tip in conformer 1. b, As in a, but for conformer 2. c, Model fitting in the FimD–tip density map. Selected densities for FimH–FimG, FimG–FimF, FimF, FimD and FimC of conformer 1 are shown. Amino acids with clear side-chain densities are indicated.
Extended Data Fig. 4 Comparison between conformer 1 and conformer 2 of the FimD–tip complex.
a, Overlap of conformer 1 (in colour cartoon) with that of conformer 2 (in grey cartoon). The movement of the NTD is labelled in the blue box, and the movement of CTD2 and FimF is labelled in the orange box. b, Comparison of FimF and CTD2 of conformer 1 with those of conformer 2. FimF and CTD2 shift 10 Å and 5 Å, respectively. c, Comparison of the NTD of conformer 1 with that of conformer 2. The NTD shifts laterally by about 30 Å and rotated by about 45°.
Extended Data Fig. 5 Comparison of interaction between plug domain and NTD of FimD and between the CTDs of FimD and FimC–FimF in conformers 1 and 2 by superimposing the two conformations.
a, Left, the plug domain and NTD of conformer 1 are coloured in salmon and orange, respectively; plug domain and NTD of conformer 2 are coloured in cyan and magenta, respectively. Right, electron densities of regions involved in the interaction in conformer I are shown for the FimD plug (top) and FimD NTD (bottom). Some amino acids have clear side-chain densities. b, Superimposition of FimD CTDs–FimC–FimF in conformer 1 (magenta) and conformer 2 (coloured as in Fig. 3). c, Detailed interactions in site 1 (marked in b). Extensive interactions are present in conformer 2. Much weaker interactions are present in conformer 1 (between Q17 and T717). d, Detailed interactions in site 2 (marked in b). In conformer 2, hydrophobic interactions exist between FimC L54 and FimD F766, and between I780 and A782 of FimD. Much weaker interactions are present in conformer 1. e, The electron densities in regions involved in interactions (in sites 1 and 2) between FimC and FimD in conformer 1. Some amino acids have side-chain densities.
Extended Data Fig. 6 Sequence conservation of the interacting interface at the two extreme termini of the FimD usher.
Residues involved in the interaction between the NTD of FimD and FimC are labelled with circles, and residues involved in the interaction between the NTD and CTD2 of FimD are labelled with triangles.
Extended Data Fig. 7 The interactions between the NTD of FimD and FimC–FimF.
a, b, The interactions between the NTD of FimD and FimC–FimF are shown for conformer 1 (a) and modelled conformer 3 (b). There are no interactions between the NTD of FimD and the FimF subunit in either conformer. The interactions between the N-terminal tail of FimD and the FimC chaperone are essentially the same in the two conformers.
Supplementary information
Video 1
Presentation of the 3D maps, segmentations, and atomic models of the FimD-tip complex in Conformers 1 and 2. The video starts by rotating the surface-rendered 3D density map of Conformer 1 around a vertical axis by 360°, then shows a still picture of the segmented density, a superimposition of the atomic model in the segmented density, followed by a 360° rotation of the atomic model shown in cartoon around the same vertical axis. The second half of the video shows the same sequence for Conformer 2: the surface-rendered 3D density, density segmentation, and atomic model shown in cartoon. The video is prepared in UCSF Chimera.
Video 2
Morph from FimD-tip Conformer 1 to Conformer 2, with an emphasis on structural changes in the NTD of FimD. The segmented 3D density of Conformer 1 is first rotated around a vertical axis by 360°. This is transitioned to the atomic model of Conformer 1 shown in cartoon. Finally, Conformer 1 is morphed into Conformer 2, and the process is repeated two times.
Rights and permissions
About this article
Cite this article
Du, M., Yuan, Z., Yu, H. et al. Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD. Nature 562, 444–447 (2018). https://doi.org/10.1038/s41586-018-0587-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0587-z
Keywords
This article is cited by
-
Stochastic chain termination in bacterial pilus assembly
Nature Communications (2023)
-
Archaic chaperone–usher pili self-secrete into superelastic zigzag springs
Nature (2022)
-
Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis
Nature Communications (2021)
-
Urinary tract infections: microbial pathogenesis, host–pathogen interactions and new treatment strategies
Nature Reviews Microbiology (2020)
-
Long polar fimbriae contribute to pathogenic Escherichia coli infection to host cells
Applied Microbiology and Biotechnology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.