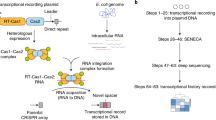
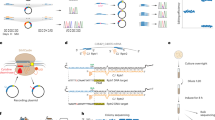
Abstract
The ability to record transcriptional events within a cell over time would help to elucidate how molecular events give rise to complex cellular behaviours and states. However, current molecular recording technologies capture only a small set of defined stimuli. Here we use CRISPR spacer acquisition to capture and convert intracellular RNAs into DNA, enabling DNA-based storage of transcriptional information. In Escherichia coli, we show that defined stimuli, such as an RNA virus or arbitrary sequences, as well as complex stimuli, such as oxidative stress, result in quantifiable transcriptional records that are stored within a population of cells. We demonstrate that the transcriptional records enable us to classify and describe complex cellular behaviours and to identify the precise genes that orchestrate differential cellular responses. In the future, CRISPR spacer acquisition-mediated recording of RNA followed by deep sequencing (Record–seq) could be used to reconstruct transcriptional histories that describe complex cell behaviours or pathological states.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Deep sequencing data are available in the National Center for Biotechnology Information Sequence Read Archive (PRJNA484149). The data sets generated and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
Karczewski, K. J. & Snyder, M. P. Integrative omics for health and disease. Nat. Rev. Genet. 19, 299–310 (2018).
Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
Ozsolak, F. & Milos, P. M. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98 (2011).
Schmidt, F. & Platt, R. J. Applications of CRISPR–Cas for synthetic biology and genetic recording. Curr. Opin. Syst. Biol. 5, 9–15 (2017).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J. & Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182 (2005).
Bolotin, A., Quinquis, B., Sorokin, A. & Ehrlich, S. D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005).
Pourcel, C., Salvignol, G. & Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 (2005).
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
van der Oost, J., Westra, E. R., Jackson, R. N. & Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 12, 479–492 (2014).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190 (2010).
Amitai, G. & Sorek, R. CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 14, 67–76 (2016).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Sternberg, S. H., Richter, H., Charpentier, E. & Qimron, U. Adaptation in CRISPR–Cas systems. Mol. Cell 61, 797–808 (2016).
Shipman, S. L., Nivala, J., Macklis, J. D. & Church, G. M. Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175 (2016).
Shipman, S. L., Nivala, J., Macklis, J. D. & Church, G. M. CRISPR–Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature 547, 345–349 (2017).
Sheth, R. U., Yim, S. S., Wu, F. L. & Wang, H. H. Multiplex recording of cellular events over time on CRISPR biological tape. Science 358, 1457–1461 (2017).
Perli, S. D., Cui, C. H. & Lu, T. K. Continuous genetic recording with self-targeting CRISPR–Cas in human cells. Science 353, aag0511 (2016).
Frieda, K. L. et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017).
Tang, W. & Liu, D. R. Rewritable multi-event analog recording in bacterial and mammalian cells. Science 360, eaap8992 (2018).
Farzadfard, F. & Lu, T. K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014).
McKenna, A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016).
Spanjaard, B. et al. Simultaneous lineage tracing and cell-type identification using CRISPR–Cas9-induced genetic scars. Nat. Biotechnol. 36, 469–473 (2018).
Kalhor, R., Mali, P. & Church, G. M. Rapidly evolving homing CRISPR barcodes. Nat. Methods 14, 195–200 (2017).
Raj, B. et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442–450 (2018).
Silas, S. et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase–Cas1 fusion protein. Science 351, aad4234 (2016).
Yosef, I., Goren, M. G. & Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 (2012).
Erdmann, S., Le Moine Bauer, S. & Garrett, R. A. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 91, 900–917 (2014).
Pyenson, N. C., Gayvert, K., Varble, A., Elemento, O. & Marraffini, L. A. Broad targeting specificity during bacterial type III CRISPR–Cas immunity constrains viral escape. Cell Host Microbe 22, 343–353 (2017).
Sandegren, L. & Sjöberg, B.-M. Self-splicing of the bacteriophage T4 group I introns requires efficient translation of the pre-mRNA in vivo and correlates with the growth state of the infected bacterium. J. Bacteriol. 189, 980–990 (2007).
Belfort, M. et al. Processing of the intron-containing thymidylate synthase (td) gene of phage T4 is at the RNA level. Cell 41, 375–382 (1985).
Gott, J. M., Shub, D. A. & Belfort, M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell 47, 81–87 (1986).
Levy, A. et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510 (2015).
Zheng, M. et al. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570 (2001).
Maurer, L. M., Yohannes, E., Bondurant, S. S., Radmacher, M. & Slonczewski, J. L. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187, 304–319 (2005).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Hardcastle, T. J. & Kelly, K. A. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11, 422 (2010).
Hassan, H. M. & Fridovich, I. Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J. Biol. Chem. 254, 10846–10852 (1979).
Ochsner, U. A., Vasil, M. L., Alsabbagh, E., Parvatiyar, K. & Hassett, D. J. Role of the Pseudomonas aeruginosa oxyR–recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB–ankB, ahpB, and ahpC–ahpF. J. Bacteriol. 182, 4533–4544 (2000).
Wesseling, C., Corriols, M. & Bravo, V. Acute pesticide poisoning and pesticide registration in Central America. Toxicol. Appl. Pharmacol. 207 (Suppl.), 697–705 (2005).
Pomposiello, P. J., Bennik, M. H. & Demple, B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183, 3890–3902 (2001).
Fuentes, D. E. et al. Cysteine metabolism-related genes and bacterial resistance to potassium tellurite. J. Bacteriol. 189, 8953–8960 (2007).
Silas, S. et al. Type III CRISPR–Cas systems can provide redundancy to counteract viral escape from type I systems. eLife 6, e27601 (2017).
Silas, S. et al. On the origin of reverse transcriptase—using CRISPR–Cas systems and their hyperdiverse, enigmatic spacer repertoires. MBio 8, e00897-17 (2017).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Biswas, A., Staals, R. H. J., Morales, S. E., Fineran, P. C. & Brown, C. M. CRISPRDetect: a flexible algorithm to define CRISPR arrays. BMC Genomics 17, 356 (2016).
Zhang, Q. & Ye, Y. Not all predicted CRISPR–Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics 18, 92 (2017).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Engler, C., Gruetzner, R., Kandzia, R. & Marillonnet, S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4, e5553 (2009).
Stead, M. B. et al. RNAsnap™: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 40, e156 (2012).
Milo, R., Jorgensen, P., Moran, U., Weber, G. & Springer, M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 38, D750–D753 (2010).
Acknowledgements
We thank M. Okoniewski for assistance with data analysis; S. Ghosh and T. Tanna for technical assistance; S. Silas, A. Z. Fire, and the entire Platt Laboratory for discussions; S. Panke, M. Jeschek, L. Pestalozzi, I. Wüthrich, and D. Gerngross for reagents and comments; C. Beisel, E. Burcklen, K. Eschbach, I. Nissen, and M. Kohler from the Genomics Facility Basel for assistance in Illumina sequencing. R.J.P., M.Y.C and F.S. are supported, in part, by funds from the Swiss National Science Foundation, ETH domain Personalized Health and Related Technologies, Brain and Behavior Research Foundation, and the National Centres of Competence – Molecular Systems Engineering.
Reviewer information
Nature thanks C. Beisel and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
F.S. and M.Y.C. performed the experiments; F.S., M.Y.C. and R.J.P. analysed the data; and F.S., M.Y.C. and R.J.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Patent applications have been filed relating to work in this manuscript.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 RT–Cas1 orthologue search and screening.
a, Experimental workflow involving the identification of 121 RT–Cas1 orthologues, overexpression in E. coli from the plasmid carrying minimal CRISPR array, containing leader–DR–spacer1–DR–spacer2–DR, followed by deep sequencing of expanded CRISPR arrays, and analysis and characterization of identified spacers. b, A comparison of the 14 disparate RT–Cas1 proteins selected for functional testing. Indicated on the left is the host species followed by a neighbour-joining phylogenetic tree built using Jukes–Cantor genetic distances of a MUSCLE multiple sequence alignment. The large ‘unknown domain’ is highlighted in green, Cas6 homology domain in pink, RT domain in purple, and Cas1 in yellow. c, Detection frequency of newly acquired spacers after overnight growth and induction of RT–Cas1–Cas2 in E. coli BL21(DE3) in different induction media. Shown is the sum of spacer counts per 1 million sequencing reads, n = 1 biological sample. d, Representative alignments of 200 spacers sequenced from F. saccharivorans array 1 to the corresponding overexpression plasmid. e, Representative alignments of 200 spacers sequenced from F. saccharivorans array 2 to the corresponding overexpression plasmid.
Extended Data Fig. 2 SENECA workflow and assessment of Record–seq efficiency in different culture conditions.
a, SENECA relies on a plasmid containing a minimal CRISPR array consisting of the leader sequence followed by a single direct repeat and a recognition sequence for the restriction enzyme FaqI. The SENECA workflows for the parental (left) and expanded (right) arrays are shown. In a Golden Gate reaction, FaqI cleaves within the direct repeat (I/II), introducing sticky ends for ligation to an Illumina P7 3′ adaptor (III). For the parental array this results in a single truncated direct repeat (IVa). For the expanded array this results in a truncated direct repeat as well as an intact direct repeat and spacer (IVb). PCR with primers binding to the full-length direct repeat and the Illumina P7 3′ adaptor results in linear amplification of the parental array (Va) and exponential amplification of the expanded array (Vb). b, Sequencing reads obtained from E. coli BL21(DE3) cells transformed with FsRT–Cas1–Cas2-encoding plasmid with or without IPTG induction. c, As in b but in E. coli BL21AI. d, As in b but in E. coli NovaBlue(DE3), a K12 substrain of E. coli. e, Percentage of sequencing reads from induced samples containing newly acquired spacers. f, Spacers per million sequencing reads obtained from cultures at an OD600 of 0.4, 0.8 or upon saturation. g, CRISPR arrays with two spacers per million sequencing reads obtained from cultures at an OD600 of 0.4, 0.8 or upon saturation. Values in b–g are mean ± s.e.m., n = 3 independent biological samples.
Extended Data Fig. 3 Record–seq-based screen of RT–Cas1 orthologues and CRISPR array directionalities.
a, Schematic of the F. saccharivorans CRISPR locus depicting the selection of CRISPR arrays and directionalities for Record–seq analysis. CRISPR arrays within each locus were identified and cloned into plasmids encoding corresponding RT–Cas1–Cas2 coding sequences. Arrays were tested in both possible directionalities, forward and reverse with a 150-bp leader. In cases of insufficient genomic data, arrays were tested in only one directionality. b, Record–seq readout of RT–Cas1 orthologues and CRISPR array directionalities. Acquisition efficiencies for forward (fw) and reverse complement (rc) directionality of each array are plotted in blue and orange, respectively. Values are genome-aligning spacers per million sequencing reads, n = 1 biological sample. n.d., not determined.
Extended Data Fig. 4 Characterization of spacers acquired by FsRT–Cas1–Cas2 and comparison of SENECA and classic spacer acquisition readouts.
a, Nucleotide probabilities determined using plasmid-aligning spacers merged across n = 14 independent biological samples, prepared as for Fig. 2f. b, Histogram of spacer GC content for all spacers or spacers acquired internal to the body of the transcript (‘gene body internal’). Values represent mean percentage of genome-aligning spacers ± s.e.m., n = 3 independent biological samples. c, Percentage of spacers aligning to either the sense or antisense strand of coding genes. The sense or antisense orientation label is with respect to the RNA, prepared as for Fig. 2c. d, Length distribution of genome-aligning spacers, prepared analogous to Fig. 2d. e, GC content distribution of genome-aligning spacers. The dotted line represents a balanced (50%) GC content, prepared as for Fig. 2e. f, Nucleotide probabilities for classic acquisition readout, prepared as for Fig. 2f. g, Nucleotide probabilities for SENECA acquisition readout, prepared analogous to Fig. 2f. Gene body coverage. For each gene the spacer coverage was determined and transformed into percentiles for comparison. Values are mean normalized coverage. n = 1 pooled sample, containing 5,798 spacers. Values in c–g are mean percentage of genome-aligning spacers, n = 1 pooled sample, containing 5,798 spacers.
Extended Data Fig. 5 Characterization of spacers acquired by FsRT–Cas1–Cas2.
a, Experimental workflow for determining the specificity of FsRT–Cas1–Cas2 for RNA using the td intron splice junction to detect RNA-derived spacers. Genomic DNA (gDNA) was extracted from an independent culture and subjected to targeted deep sequencing of the td intron insertion site. b, Quantification of td intron splice junctions. The splice junction is specific to RNA-derived spacers and not genomic DNA or cDNA copies generated by alternative reverse transcriptases in the E. coli genome. Values represent mean td intron splice junction counts per million sequencing reads ± s.e.m., n = 3 independent biological samples. c, Number of spacers aligned to plasmid, E. coli genome, and MS2 genome, showing CRISPR acquisition from an RNA virus. The total number and percentage of spacers aligning to each reference are shown. Values represent the sum of MS2-aligning spacers across replicates, n = 64 technical replicates from n = 2 biological samples, representing 22 million spacers. d, Number of MS2-aligned spacers from c that align to the overexpression plasmid, E. coli and MS2 genome, showing that MS2-aligned spacers are specific to the MS2 genome. The total number and percentage of MS2-aligned spacers that subsequently align to each reference are shown, n = 64 technical replicates from n = 2 biological samples, representing 22 million spacers. e, Total number of spacers aligning to features of the MS2 genome, n = 64 technical replicates from n = 2 biological samples, representing 22 million spacers. f, Scatter plot of transcript counts from the MS2 and E. coli genomes. Each dot represents the mean spacer count for each transcript, n = 4 independent biological samples. The horizontal black bars are mean genome-aligning spacer count across all transcripts ± s.e.m.
Extended Data Fig. 6 Quantitative analysis of arbitrary RNA sequence recording using qRT–PCR and Record–seq.
a, Coverage of spacers from Fig. 3f aligning to sfGFP or Rluc. Arrow and dotted line reflect the transcription start site (TSS), black octagon indicates the transcriptional terminator. For each nucleotide position, the sum spacer coverage per million sequencing reads is shown, n = 10 independent biological samples. b, Absolute quantification of sfGFP mRNA measured by qRT–PCR. Samples from Fig. 3f. Values are mean ± s.e.m. copy number per 6 × 109 cells, normalized by 16S rRNA copy number, n = 10 independent biological samples. c, As in b, but for Rluc. d, Scatter plot depicting the correlation between absolute sfGFP mRNA copy number and the number of transcript-aligning spacers from Fig. 3f. Linear regression fit, coefficient of determination (R2), and Pearson linear correlation coefficient (P), n = 10 independent biological samples. e, As in d, but for Rluc. f, Comparison of spacer counts for arbitrary sfGFP sequence and endogenous transcripts. Each dot represents the mean spacer count for each transcript, horizontal black bars are mean genome-aligning spacer count ± s.e.m., n = 10 independent biological samples. g, Dose–response relationship between sfGFP-aligning spacers and inducer concentration for different numbers of recorded spacers. These data represent the average number of sfGFP-aligning spacers ± s.e.m., n = 10 independent biological samples. h, Relative spacer count of spacers mapping to the Fluc transcript after 3OC6-HSL induction. Values are the normalized mean number of spacers per million sequencing reads ± s.e.m. with n = 6 independent biological samples. i, Absolute quantification of Fluc mRNA measured by qRT–PCR. Data were obtained from the same bacterial cultures as in Fig. 3g. Values are mean copy number per 6 × 109 cells, normalized by 16S rRNA copy number, ± s.e.m., n = 10 independent biological samples. j, As in i, but for Rluc.
Extended Data Fig. 7 Record–seq reveals cumulatively highly expressed genes.
a, Scatter plots depicting Record–seq correlation between n = 3 independent biological replicates shown in b and c. Linear regression fit, coefficient of determination (R2), and Pearson linear correlation coefficient (P) are shown for each comparison. Data represent log2-normalized transcript quantification counts. b, Spacers are preferentially acquired from highly expressed genes. Record–seq spacer counts for plasmid and E. coli genes (top) or only E. coli genes (bottom) according to decreasing RNA-seq-based gene expression values. Monte Carlo bounds reflect simulated spacers with no transcriptional bias. Mean cumulative normalized spacer count, and Monte Carlo bounds are shown, n = 3 independent biological samples. c, Assessing the correlation between an RNA-seq stationary phase snapshot and a Record–seq transcriptional record. RNA-seq and Record–seq were performed on the same population of E. coli BL21(DE3) in stationary phase growth, induced to express FsRT–Cas1–Cas2 overnight. The correlation between all (top left), stationary-phase (top right), log-phase (bottom left), and plasmid-borne (bottom right) genes are shown. The linear regression fit, coefficient of determination (R2), and Pearson linear correlation coefficient (P) are shown for each comparison. The data represent the log2 normalized transcript quantification counts averaged across replicates, n = 3 independent biological samples. d, Correlation of Record–seq with log- and stationary-phase genes over long-term cultivation. These data represent the R2 value calculated as described for b for either stationary or logarithmic phase gene sets using different E. coli culture time points as inputs with n = 3 independent biological samples. e, Comparison of transcript-aligning spacer counts with and without normalizing for gene expression level. Each dot represents the mean normalized number of counts per transcript with n = 3 independent biological samples. The horizontal black bars are mean genome-aligning spacer count ± s.e.m. See Supplementary Notes for detailed discussions on b, d.
Extended Data Fig. 8 Defining the minimum number of cells required for assessing complex cellular behaviours using Record–seq and PCA.
Using the acid stress response data set shown in Fig. 4, PCA was performed on the entire data set as well as progressively and randomly downsampled data. These data show that Record–seq appropriately classifies the acid stress response samples with 7% of the original data (corresponding to 314 spacer or 6.1 × 106 E. coli cells). The calculation of the required number of E. coli cells is described in detail in the Supplementary Notes; n = 10 independent biological samples.
Extended Data Fig. 9 Defining the minimum number of cells required for assessing complex cellular behaviours using Record–seq and differential expressed signature gene analysis.
Using the acid stress response data set shown in Fig. 4e–g, differential expressed signature genes were identified for the entire data set as well as progressively and randomly downsampled data. The plots depict hierarchically clustered signature gene heatmaps. These data show that with 10% of the original data (corresponding to 448 spacer or 8.8 × 106 E. coli cells) the signature genes can appropriately classify the samples. The calculation of cell numbers is described in detail in the Supplementary Notes; n = 10 independent biological samples.
Extended Data Fig. 10 Optimization of CRISPR spacer acquisition efficiency and detection of signature genes corresponding to Record–seq-compatible sentinel cells for encoding transient herbicide exposure.
a, Plasmid and genome-aligning spacers obtained from E. coli BL21(DE3) transformed with FsRT–Cas1–Cas2 encoding plasmid using the original coding sequence (CDS) (light blue) or optimized CDS (dark blue) under the indicated IPTG concentrations. b, Plasmid and genome-aligning spacers obtained from E.coli BL21(DE3) transformed with FsRT–Cas1-Cas2 encoding plasmid using the optimized coding sequence under transcriptional control of the PT7lac, PtetA, or PrhaB promoter, induced with the indicated concentrations of IPTG, aTc, or Rhamnose, respectively. c, Unsupervised hierarchical clustering of RNA-seq cumulative expression profiles for signature differentially (cumulatively) expressed genes. Signature genes represent the union between the top 20 most differently expressed genes identified by DESeq2, edgeR, and baySeq, n = 6 independent biological samples. d, Unsupervised hierarchical clustering of Record–seq cumulative expression profiles for signature differentially (cumulatively) expressed genes. Signature genes represent the union between the top 20 most differently expressed genes identified by DESeq2, edgeR, and baySeq, n = 6 independent biological samples. Data in a, b are mean ± s.e.m., n = 3 independent biological samples.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Primer Design Notes 1-2, Supplementary Tables 1-9 and Supplementary Sequences 1-3
Supplementary Table
This file contains Supplementary Table 2.
Supplementary Table
This file contains Supplementary Table 9.
Rights and permissions
About this article
Cite this article
Schmidt, F., Cherepkova, M.Y. & Platt, R.J. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature 562, 380–385 (2018). https://doi.org/10.1038/s41586-018-0569-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0569-1
Keywords
This article is cited by
-
Synthetic CRISPR/dCas9-KRAB system driven by specific PSA promoter suppresses malignant biological behavior of prostate cancer cells through negative feedback inhibition of PSA expression
Cellular & Molecular Biology Letters (2023)
-
Histones direct site-specific CRISPR spacer acquisition in model archaeon
Nature Microbiology (2023)
-
Retrospective analysis of enhancer activity and transcriptome history
Nature Biotechnology (2023)
-
Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays
Nature Structural & Molecular Biology (2023)
-
RNA recording in single bacterial cells using reprogrammed tracrRNAs
Nature Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.