Abstract
Defects in the architecture or integrity of the nuclear envelope are associated with a variety of human diseases1. Micronuclei, one common nuclear aberration, are an origin for chromothripsis2, a catastrophic mutational process that is commonly observed in cancer3,4,5. Chromothripsis occurs after micronuclei spontaneously lose nuclear envelope integrity, which generates chromosome fragmentation6. Disruption of the nuclear envelope exposes DNA to the cytoplasm and initiates innate immune proinflammatory signalling7. Despite its importance, the basis of the fragility of the micronucleus nuclear envelope is not known. Here we show that micronuclei undergo defective nuclear envelope assembly. Only ‘core’ nuclear envelope proteins8,9 assemble efficiently on lagging chromosomes, whereas ‘non-core’ nuclear envelope proteins8,9, including nuclear pore complexes (NPCs), do not. Consequently, micronuclei fail to properly import key proteins that are necessary for the integrity of the nuclear envelope and genome. We show that spindle microtubules block assembly of NPCs and other non-core nuclear envelope proteins on lagging chromosomes, causing an irreversible defect in nuclear envelope assembly. Accordingly, experimental manipulations that position missegregated chromosomes away from the spindle correct defective nuclear envelope assembly, prevent spontaneous nuclear envelope disruption, and suppress DNA damage in micronuclei. Thus, during mitotic exit in metazoan cells, chromosome segregation and nuclear envelope assembly are only loosely coordinated by the timing of mitotic spindle disassembly. The absence of precise checkpoint controls may explain why errors during mitotic exit are frequent and often trigger catastrophic genome rearrangements4,5.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. Source data for all graphs (Figs. 1c, 2a, b, 3a, c, 4b, d, e; Extended Data Figs. 1d, f, 2b, c, d, f, g, 3a–c, e, 4c, d, f, g, 5b, c, e, 6a–c, 7b, d, e, 8d, 9a, d–f, h, 10c, d) are provided with the online version of the paper. All other data sets generated and/or analysed in the current study are available from the corresponding author upon reasonable request. Supplementary Fig. 1 contains scanned complete images of western blots.
References
Ungricht, R. & Kutay, U. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 18, 229–245 (2017).
Zhang, C.-Z. et al. Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Notta, F. et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 538, 378–382 (2016).
Ly, P. & Cleveland, D. W. Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol. 27, 917–930 (2017).
Hatch, E. M., Fischer, A. H., Deerinck, T. J. & Hetzer, M. W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 (2013).
Umbreit, N. T. & Pellman, D. Cancer biology: Genome jail-break triggers lockdown. Nature 550, 340–341 (2017).
Haraguchi, T. et al. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 121, 2540–2554 (2008).
Dechat, T. et al. LAP2α and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 117, 6117–6128 (2004).
Otsuka, S. et al. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife 5, e19071 (2016).
de Castro, I. J., Gil, R. S., Ligammari, L., Di Giacinto, M. L. & Vagnarelli, P. CDK1 and PLK1 coordinate the disassembly and reassembly of the nuclear envelope in vertebrate mitosis. Oncotarget 9, 7763–7773 (2017).
Toledo, L. I. et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013).
Hatch, E. M. & Hetzer, M. W. Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol. 215, 27–36 (2016).
Crasta, K. et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012).
Afonso, O. et al. Feedback control of chromosome separation by a midzone aurora B gradient. Science 345, 332–336 (2014).
Karg, T., Warecki, B. & Sullivan, W. Aurora B-mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol. Biol. Cell 26, 2227–2241 (2015).
Fuller, B. G. et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature 453, 1132–1136 (2008).
Ramadan, K. et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase aurora B from chromatin. Nature 450, 1258–1262 (2007).
Vagnarelli, P. et al. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 21, 328–342 (2011).
Obara, Y., Chai, L. S., Weinfeld, H. & Sandberg, A. A. Synchronization of events in fused interphase-metaphase binucleate cells: progression of the telophase-like nucleus. J. Natl. Cancer Inst. 53, 247–259 (1974).
Walther, T. C. et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113, 195–206 (2003).
Xue, J. Z., Woo, E. M., Postow, L., Chait, B. T. & Funabiki, H. Chromatin-bound Xenopus Dppa2 shapes the nucleus by locally inhibiting microtubule assembly. Dev. Cell 27, 47–59 (2013).
Vietri, M. et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231–235 (2015).
Hochegger, H., Hégarat, N. & Pereira-Leal, J. B. Aurora at the pole and equator: overlapping functions of aurora kinases in the mitotic spindle. Open Biol. 3, 120185 (2013).
Lu, L., Ladinsky, M. S. & Kirchhausen, T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J. Cell Biol. 194, 425–440 (2011).
Kurasawa, Y., Earnshaw, W. C., Mochizuki, Y., Dohmae, N. & Todokoro, K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237–3248 (2004).
Barisic, M. et al. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803 (2015).
Otsuka, S. et al. Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat. Struct. Mol. Biol. 25, 21–28 (2018).
Poser, I. et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 (2008).
Gruneberg, U., Neef, R., Honda, R., Nigg, E. A. & Barr, F. A. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166, 167–172 (2004).
Soto, M. et al. p53 prohibits propagation of chromosome segregation errors that produce structural aneuploidies. Cell Reports 19, 2423–2431 (2017).
Rieder, C. L. & Cassels, G. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods Cell Biol. 61, 297–315 (1999).
Hiraoka, Y., Sedat, J. W. & Agard, D. A. Determination of three-dimensional imaging properties of a light microscope system. Partial confocal behavior in epifluorescence microscopy. Biophys. J. 57, 325–333 (1990).
Gustafsson, M. G. et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 (2008).
Belgareh, N. et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154, 1147–1160 (2001). https://doi.org/10.1083/jcb.200101081.
Maeshima, K. et al. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J. Cell Sci. 119, 4442–4451 (2006).
Clever, M., Funakoshi, T., Mimura, Y., Takagi, M. & Imamoto, N. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus 3, 187–199 (2012).
Mimura, Y., Takagi, M., Clever, M. & Imamoto, N. ELYS regulates the localization of LBR by modulating its phosphorylation state. J. Cell Sci. 129, 4200–4212 (2016).
Hudson, D. F., Vagnarelli, P., Gassmann, R. & Earnshaw, W. C. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell 5, 323–336 (2003).
Maiato, H., Afonso, O. & Matos, I. A chromosome separation checkpoint: A midzone Aurora B gradient mediates a chromosome separation checkpoint that regulates the anaphase-telophase transition. BioEssays 37, 257–266 (2015).
Maciejowski, J., Li, Y., Bosco, N., Campbell, P. J. & de Lange, T. Chromothripsis and kataegis induced by telomere crisis. Cell 163, 1641–1654 (2015).
Denais, C. M. et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016).
Steigemann, P. et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136, 473–484 (2009).
Uehara, R. et al. Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J. Cell Biol. 202, 623–636 (2013).
Sagona, A. P., Nezis, I. P. & Stenmark, H. Association of CHMP4B and autophagy with micronuclei: implications for cataract formation. BioMed Res. Int. 2014, 974393 (2014).
Santaguida, S. et al. Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651 (2017).
Shah, P., Wolf, K. & Lammerding, J. Bursting the bubble—nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. 27, 546–555 (2017).
Acknowledgements
We thank I. Cheeseman, T. Rapoport, N. Umbreit, T. Walther, and K. Xie for advice or comments on the manuscript; E. Jackson and A. Spektor for preliminary experiments; R. Davidowitz for animations; J. Ellenberg, D. Gerlich, E. Hatch, M. Hetzer, A. Hyman, T. Kuroda, and L. Shao for reagents; and J. Waters and T. Lambert of the Nikon Imaging Center at Harvard Medical School and H. Elliott and C. Yapp from the Image and Data Analysis Core at Harvard Medical School for advice. We acknowledge the use of the Wadsworth Center’s Electron Microscopy Core Facility. A. K. is supported by NIH GM059363. D.P. is a HHMI investigator and is supported by R37 GM61345.
Author information
Authors and Affiliations
Contributions
D.P., S.L., and M.K. designed the experiments. D.P., S.L., and M.K. wrote the manuscript, with edits from all authors. S.L. and M.K. performed most experiments and analysis. M.M. assisted with several experiments and contributed Extended Data Figs. 4g, 5b, 8d, 10c, d. N.Y. and A.K. performed the electron microscopy in Extended Data Fig. 5g. F.R. and A.K. performed the electron microscopy and contributed Fig. 2b.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
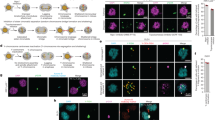
Extended Data Fig. 1 Defective NE assembly on lagging chromosomes (or chromosome bridges).
a, Cartoon of NE subdomains that form transiently around the main chromosome mass in a telophase cell8,9 and summarizing results for lagging chromosomes. DNA, light blue. BAF, barrier-to-autointegration factor. The NPC is a complex containing NUPs, including NUP133, NUP107, ELYS, TPR, NUP153, NUP358 and NUP621 (see d and Extended Data Fig. 2f). b, Live-cell/fixed cell imaging showing the recruitment of core (emerin) and non-core (NUP133) proteins onto lagging chromosomes (yellow arrows) and the main chromosome mass at the indicated times after anaphase onset (AO, t = 0). Top, experimental scheme. RFP–H2B-expressing RPE-1 cells were released from the nocodazole mitotic block, imaged on gridded coverslips at 2-min intervals, and then fixed and labelled for immunofluorescence (Extended Data Fig. 4e). Bottom, cartoons (left) and images of RPE-1 cells (right) showing DNA (blue), emerin (red) and NUP133 (green). Each image represents 20 cells (from two technical replicates) from the indicated time points. Scale bars, 10 µm. Note there is some variability between cells (~1–2 min) in the times at which different proteins are recruited. During early–mid anaphase (2–4 min post AO), NUP133 is on kinetochores35 (green dots in cartoon). About 4–6 min after AO, NUP133 begins to assemble on the chromosome periphery (right-most column, white arrowheads). About 6–8 min after AO, emerin assembles on lagging chromosome and the main chromosome mass, including the region adjacent to the central spindle (right-most column, orange arrowheads, microtubules not shown). By 8–12 min after AO, emerin becomes concentrated in a recognizable core domain8, which is also detected as a gap in NUP133 signal (enlarged image). The peripheral localization of NUP133 (for example, 8 min) marks the non-core domain. About half of the NPCs of the interphase nucleus assemble in this ~8–10-min period of telophase28 (hereafter referred to as late mitotic NPC assembly). Nuclear pore proteins display the most obvious non-core gap, whereas other non-core proteins, such as LBR, more commonly display reduced signal intensity within the core domain (e, top row, RPE-1). Defective NUP133 assembly on lagging chromosomes persists throughout mitotic exit (>15 min after AO). By ~15 min after AO, the core and non-core domains become intermingled on the main nucleus, with fragments of the core domain persisting as pore-free islands that are slowly populated by NPCs during interphase10,36. c, Similar to RPE-1 cells (b), images of HeLa K cells (representative of 30 cells, from two experiments) showing that the core membrane proteins emerin (top two rows) and LAP2β (bottom two rows) first associate with the chromosome periphery (yellow arrowheads) contemporaneously with the non-core (NPC, mAb414 detects FG-containing nucleoporins) proteins. About 2–4 min later, the core proteins extend into and then concentrate in (emerin, orange arrowheads; LAP2β does not concentrate) the core domain (red arrowheads). Cells were synchronized as in e. In HeLa K cells, lagging chromosomes often exhibit a slight delay (~1–2 min) in the recruitment of core membrane proteins (emerin and LAP2β) as compared to the periphery of the main chromosome mass. Scale bars, 10 µm. It is thought that the NE assembles from a continuous network of mitotic endoplasmic reticulum (ER)1. It is therefore simplest to propose that the core and non-core subdomains are also in a continuous network, but the core domain is just a region of the continuous ER network that is missing the non-core subgroup of proteins. Supporting this idea, prior work8 and our data (b, c) show that the core proteins initially assemble together with the non-core proteins around the chromosome periphery and only later become enriched near the microtubules. This data suggests that the domain partitioning could come solely from NPC precursors and LBR (which requires ELYS for recruitment37,38) being preferentially retained in fenestrated ER sheets28 that might less readily penetrate bundled spindle microtubules (Fig. 3a). Although we favour this model, we cannot exclude the possibility that core and non-core proteins somehow partition into separate membrane compartments. d, Quantification of defective non-core NE protein recruitment to lagging chromosomes. Synchronization as in Fig. 1a (n = 64, 118, 151, 124, 151, 145, 150, 69, 90, 65, 70, 60, 64, left to right, from three experiments for RPE-1 cells, n = 149, 110, 124, 132, from two experiments for HeLa K cells and n = 44, 76, from two experiments for U2OS cells). e, f, Orthogonal method (1 µM NMS-P715, MPS1i) to generate lagging chromosomes shows a similar non-core NE assembly defect to the nocodazole block-and-release protocol (Fig. 1a). e, Top, experimental scheme. Bottom, representative images of RPE-1 and HeLa K cells. In RPE-1 and U2OS cells there is a near absence of non-core protein on lagging chromosomes, regardless of the method of generation. In HeLa K cells, the effect is less penetrant. About 60% of lagging chromosomes lack detectable non-core protein recruitment, ~15% display strongly reduced levels (scored as labelled but shown in grey bar as reduced, f), and ~25% display a clear signal (NUP133), but often only covering part of the circumference of the lagging chromosome. These differences are probably due to differences in spindle organization between cell lines (Fig. 3; Extended Data Figs. 6–8). f, Quantification of the results (n = 56, 78 for RPE-1 cells, from two experiments; n = 75, 174 for HeLa K cells, from three experiments). Scale bars, 10 µm. g, h, Chromatin bridges (arrows) formed after nocodazole release or partial depletion of SMC239 show core-only NE protein assembly. g, An RPE-1 cell after release from a nocodazole block (representative of 30 DNA bridges from five experiments). h, Top, experimental scheme for generating chromosome bridges by partial SMC2 depletion. Bottom, an emerin–GFP-expressing RPE-1 cell. Percentages of cells with the indicated staining pattern are shown (n = 30, from two experiments). Scale bars, 10 µm. DNA bridges are uniformly depleted for non-core (LBR) proteins, with no evidence for a gradient as might be expected for the chromosome separation checkpoint hypothesis15,40. Interphase chromatin bridges have been reported to have an altered NE protein composition, including reduced levels of lamin B1 and NPCs41, which is consistent with our findings.
Extended Data Fig. 2 Defect in NPC and other non-core protein assembly persists in interphase MN in multiple cell lines.
a–c, Defective non-core NE protein recruitment to MN in RPE-1 cells. a, Top, experimental scheme. Bottom, representative images (quantified in b) with MN (arrows in enlarged images). Red letters, core NE proteins; green, non-core proteins. b, Quantification of results from a. The FI ratio of the indicated NE proteins in intact MN (RFP–NLS positive) relative to PN at the indicated time points after release from nocodazole block (mean with 95% CI, n = 97, 120, 106, 92, 114, 104, 113, 116, 104, 114, 121, 91, 114, 89, 113, 116, 125, left to right, from two experiments). Scale bars, 10 µm. c, MN/PN FI ratio of LAP2β in RPE-1 cells at the indicated time points after release from nocodazole block (mean with 95% CI, n = 70, 49, from two experiments). d, Deficiency of non-core proteins persists in MN in HeLa K (left) and U2OS cells (right). MN/PN FI ratio of the indicated NE proteins after release from nocodazole block as in a (mean with 95% CI, n = 79, 84, 111, 90, 79, 84, 111, 90, left to right for HeLa K, from two experiments; n = 53, 47, 45, 53, 66, 23, 64, 53, 27, 66, 54, 28, 53, 47, 46, 53, 69, 23, left to right for U2OS, from two experiments). e, Representative images of U2OS cells from d showing reduced assembly of B-type lamins in MN (arrows) 90 min after nocodazole release. Scale bars, 10 µm. f, Reduced NPC assembly on MN. MN/PN FI ratio of NPC proteins in RPE-1 cells 15 h after release from nocodazole block (mean with 95% CI, n = 47, 44, 49, 51, 49, 56, left to right, from two experiments). g, Reduced accumulation of B-type lamins but not A-type lamins on intact MN. Left, representative images of RPE-1 (left) and HeLa K (right) cells. In the enlarged merged images from the orange boxed region (HeLa K), the intensity of lamin B1 has been scaled differently to illustrate two points. First, there is reduction of lamin B but not lamin A/C in MN, which becomes evident when lamin B and lamin A/C on the main nucleus are scaled to the same intensity. Second, some MN display a lamin B1 gap, as has been reported6 (arrowheads indicate lamin B1 gap formed where the lamin A/C rim is continuous), which becomes evident when the lamin B1 intensity is scaled to a higher level. Right, general reduction of B-type lamins in MN. MN/PN FI ratio for the indicated lamins. Shown is the background-subtracted raw data for each individual micronucleus analysed in the indicated cell lines (n = 116 for RPE-1, n = 111 for HeLa K, from b, d above), 5 h after nocodazole release. Scale bars, 10 µm. A fraction of MN exhibit reduced lamin A/C, possibly owing to impaired import of lamin A/C during interphase.
Extended Data Fig. 3 MN have defective nuclear import and impaired accumulation of nuclear proteins.
a, b, Defective nuclear import kinetics in newly formed MN visualized by high temporal resolution, confocal live-cell imaging. MN were generated in RPE-1 (a) or U2OS (b) cells as in Fig. 1a. a, Defective nuclear RFP–NLS import in RPE-1 cells. Left, categories of cells with differing levels of accumulation of the import reporter in MN as compared to PN (see Fig. 1c for quantification). Middle, nuclear import kinetics from representative RPE-1 cells expressing GFP–H2B and RFP–NLS for PN (blue) and MN (red). Black lines and shaded regions indicate mean ± s.d. from 11 PN. To control for photobleaching, the bleaching coefficient of the RFP–NLS signal was experimentally determined by imaging of cells during late interphase when nuclear import was presumed to be at steady state. A correction based on this bleaching coefficient obtained for 12 PN/MN pairs was then applied to the data presented here (see Methods). Right, representative images from a time-lapse series from RPE-1 cells expressing GFP–H2B (left) and RFP–NLS (right). Arrows indicate a pair of PN with RFP–NLS import evident from 10 min. Yellow boxes show lagging chromosomes and the resulting MN with nuclear RFP–NLS accumulation defects (insets, enlarged images). Bottom right, heat map of RFP–NLS intensity (t = 53 min) illustrating the strong import defect in MN (see Supplementary Videos 1, 2). b, Defective nuclear GFP–IBB import in lagging chromosomes from U2OS cells. Top, percentage of MN corresponding to the categories in a in U2OS cells expressing RFP–H2B and GFP–IBB (n = 19, from 17 experiments). Bottom, images from a time-lapse series (boxes, lagging chromosomes and MN). Scale bars, 10 µm. c, Impaired nuclear accumulation of multiple proteins in MN relative to PN. Top, representative images of RPE-1 cells with intact or disrupted MN (arrows). Micronucleus disruption is visualized by hyper-accumulation of GFP–BAF42. Both intact and disrupted MN lack detectable accumulation of RPA2. Middle, MN/PN FI ratio of RPA2 (mean with 95% CI, n = 62, 23, from two technical replicates). MN were generated in RPE-1 cells as in Extended Data Fig. 1e. Bottom, FI ratio of indicated proteins in intact (RFP–NLS positive) MN from RPE-1 cells at indicated time points after nocodazole release as in Extended Data Fig. 2a (mean with 95% CI, n = 89, 98, 89, 98, 107, 100, 107, 100, left to right, from three experiments). LSD1, lysine-specific histone demethylase 1; Rb, retinoblastoma protein. Scale bars, 10 µm. d, e, Live-cell/fixed cell imaging showing impaired nuclear accumulation of RPA2 and RFP–NLS in MN after mitotic exit. d, Top, experimental scheme. Bottom, images from wide-field time-lapse series followed by fixation and confocal imaging. Confocal images (from red and blue boxed regions) illustrate reduced accumulation of RFP–NLS and RPA2 in newly formed MN (yellow and red arrows, respectively). Time after anaphase onset (t = 0 min) is shown. Scale bars, 10 µm. e, Top, MN/PN FI ratio of RPA2 and RFP–NLS in RPE-1 cells ~1 h after anaphase onset (mean with 95% CI, n = 211, from two experiments). Bottom, defective accumulation of RPA2 and RFP–NLS are correlated (r = 0.9039, P < 0.0001, two-tailed Spearman’s correlation).
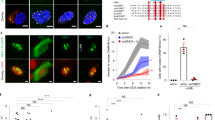
Extended Data Fig. 4 Proximity to spindle midzone Aurora B does not affect the NE assembly defect of lagging chromosomes; this defect persists through late telophase regardless of whether Aurora B is active.
a, Core membrane proteins (LAP2β and emerin) assemble on lagging chromosomes and chromosome bridges, independent of their position relative to the spindle midzone. Top, images of an RPE-1 cell expressing GFP–BAF and RFP–LAP2β show recruitment of LAP2β to lagging chromosome (arrow). Bottom, images of an RPE-1 cell expressing emerin–GFP show recruitment of emerin to a chromosome bridge (arrowhead). Both LAP2β and emerin can assemble near the concentration of aurora B (white, in merged images) at the spindle midzone (representative of 30 cells, from two experiments). Cells synchronized as in Fig. 1a. Scale bars, 10 µm. b, c, Live-cell/fixed cell imaging shows that loss of H3S10 phosphorylation in late telophase does not enable non-core NE assembly onto lagging chromosomes. b, Top, experimental scheme. Bottom, representative images of RPE-1 cells. Scale bars, 10 µm. c, FI measurements from the main chromosome mass (PN) and lagging chromosome (MN) showing loss of H3S10 phosphorylation relative to the recruitment of NPC proteins in the same cell at indicated time points. Top, phospho-H3S10 with a mouse monoclonal anti-pH3S10 antibody (mean with 95% CI, n = 31, 46, 34, 27, 26, 28, 34, 26 each time point for MN and PN mouse-pH3S10, from two experiments; n = 31, 46, 34, 27, 26, 28, 34, 26 each time point for MN and PN NUP133, from two experiments). Bottom: rabbit polyclonal anti-pH3S10 antibody (mean with 95% CI, n = 27, 20, 33, 29, 22, 30, 22, 32, 31, 27 each time point for MN and PN rabbit-pH3S10, from two experiments; n = 30, 49, 28, 32, 30, 33, 32, 32, 27 each time point for MN and PN NUP107, from two experiments). Zero time point measurements for both MN and PN were from metaphase chromosomes. The specificity of both anti-pH3S10 antibodies was confirmed by the near-complete loss of labelling after treatment of nocodazole-arrested mitotic cells with 50 µM ZM447439 (not shown). Note that NUP133 starts to assemble on the main chromosomes (~4–6 min after anaphase onset28, also see Extended Data Fig. 5b) with readily detected H3S10 phosphorylation. d, MKLP2 knockdown, disrupting the transport of aurora B to the spindle midzone, fails to restore non-core (LBR) NE assembly to lagging chromosomes (arrows). Synchronization as in Fig. 1a. Left, representative images of control or MKLP2-depleted RPE-1 cells. Middle, MN/PN FI ratio of LBR. For MN in MKLP2-depleted cells, only cells lacking detectable midzone aurora B were quantified (mean with 95% CI, n = 118, 105, from three experiments). NS, P = 0.8147, two-tailed Mann–Whitney test. Right, western blot showing MKLP2 knockdown in RPE-1 cells (two experiments, for gel source data see Supplementary Fig. 1). Scale bars, 10 µm. e, Live-cell/fixed cell imaging protocol. RPE-1 cells expressing RFP–H2B were plated on gridded dishes to identify cells of interest. Cells were imaged at 2-min intervals. Representative images of two live cells (red and blue boxes) upon (left) or after (right, before fixation) ZM447439 addition. f, Consistent with prior studies17,43,44, aurora B inhibition (ZM447439, 5 µM or 50 µM) rapidly (3 min) inactivates the kinase (mean with 95% CI, n = 48, 17, 19, from two technical replicates). ****P < 0.0001, two-tailed Mann–Whitney test. Active aurora B was assessed by the FI ratio of phospho-aurora B (pT232) to total aurora B in the midbody of telophase cells 3 min after addition of ZM447439 or DMSO. RPE-1 cells were synchronized as in Fig. 1a. g, Tenfold higher (50 µM) concentration of aurora B inhibitor gives similar results to those obtained with 5 µM inhibitor (Fig. 2a), confirming that early aurora B inhibition (2 min before to 2 min after anaphase onset) but not late (6–12 min after anaphase onset) partially restores non-core protein (NUP133) recruitment to lagging chromosomes. Note that non-core NE is only partially restored on a fraction of lagging chromosomes even with the high dose of inhibitor (early 50 µM ZM447439) even though this treatment led to a uniform loss of phospho-H3S10 on all chromosomes after anaphase entry (not shown). Therefore, the persistent NE assembly defect on lagging chromosomes after aurora B inhibition cannot be explained by residual phospho-H3S10. Instead, the partial effect is explained by the effects of aurora B inhibition on the local organization of microtubules (Extended Data Figs. 6a, 8d). Shown is the MN/PN FI ratio of NUP133 at the indicated time intervals and drug doses (mean with 95% CI, n = 33, 37, 37, 52, 37, left to right, from two experiments). Experiments performed as in Fig. 2a in RPE-1 cells. For simplicity, time points corresponding to ± 2 min after anaphase onset were grouped as early treatment, whereas time points corresponding to 6–12 min after anaphase onset were grouped as late treatment. ***P = 0.0001 (DMSO and early 5 µM ZM), ***P = 0.0003 (early 5 µM ZM and late 5 µM ZM), ****P < 0.0001; NS, P = 0.494, two-tailed Mann–Whitney test.
Extended Data Fig. 5 The irreversible NE assembly defect of lagging chromosomes may be caused by enclosure in an NPC-deficient NE, rather than persistent condensins.
a–c, Loss of condensin (SMC2) is not required for non-core and NPC assembly. a, Representative images of control and SMC2-depleted RPE-1 cells showing that loss of SMC2 does not restore non-core assembly on lagging chromosomes (quantified in b, c). Scale bar, 10 µm. b, Top, experimental scheme of live-cell/fixed cell imaging. Middle and bottom, FI of NUP133 and SMC2 on the main chromosome mass (PN) and lagging chromosomes (MN) at time points after anaphase onset in RPE-1 cells (mean with 95% CI, n = 52, 37, 35, 44, 35, 36, 32, 46, 41 each timpoint for MN SMC2, from three experiments; n = 52, 37, 47, 55, 40, 48, 38, 46, 41 each time point for PN SMC2, from three experiments; n = 31, 28, 37, 22, 23, 25, 26, 28, 34 each time point for MN and PN NUP133, from two experiments). Zero time point measurements are from metaphase chromosomes. Dashed lines indicate time points, demonstrating that NUP133 can assemble on the PN when condensins are present at ~70% of their maximum levels in metaphase. Note that for MN (~10 min after anaphase onset), the decline in SMC2 levels is slower than on the PN, presumably because of nuclear transport is required for complete chromosome decondensation. c, Top, SMC2 levels on PN and MN during telophase in control and SMC2-depleted cells (mean with 95% CI, n = 58, 58, 75, 75, left to right, from two experiments). ****P < 0.0001, two-tailed Mann–Whitney test. Middle, MN/PN FI ratio of mAb414 to detect nucleoporins. For MN in SMC2-depleted cells, only cells with no detectable SMC2 were quantified for mAb414 (mean with 95% CI, n = 35, 45, from two experiments). NS, P = 0.9251, two-tailed Mann–Whitney test. Bottom, western blot showing SMC2 knockdown in RPE-1 cells (two experiments; for gel source data see Supplementary Fig. 1). d, e, Recruitment of ESCRT-III to lagging chromosomes. d, Normal kinetics for the association and dissociation of ESCRT-III (CHMP4B–GFP) on lagging chromosomes (white arrowheads) during NE reformation. Lagging chromosomes were induced in HeLa K cells expressing CHMP4B–GFP (green) and RFP–H2B (red) by nocodazole block-and-release as in Fig. 1a and imaged at 1-min intervals on a confocal microscope (Supplementary Video 3). Left, a representative cell showing the CHMP4B residence time on lagging chromosome (MN) as compared to the residence time on the PN. Right, enlarged images of boxed regions showing lagging chromosome (white arrowheads). The duration of CHMP4B association with the PN or MN is shown by the coloured bars above the time-lapse series. Similar results were obtained for chromosome bridges (not shown). Note that prior work suggests that the timely dissociation of CHMP4B is a marker for successful NE sealing and that delayed CHMP4B dissociation is a marker for defective NE sealing23. The normal ESCRT-III kinetics on lagging chromosomes or chromosome bridges therefore suggests substantial NE membrane sealing on these structures (representative of ten cells from six experiments). ESCRT-III was not detected on newly formed MN after mitotic exit (not shown). ESCRT-III has been reported to associate with interphase MN45, which we also observe, but only on disrupted MN (not shown). In addition, the previously described23 enrichment of ESCRT-III in the core domain of the main chromosome mass is less apparent after nocodazole release (shown here) than is observed when lagging chromosomes are generated by MPS1i treatment (Extended Data Fig. 9b). Scale bars, 10 µm. e, ESCRT-III (CHMP4B, CHMP2A and IST1) is recruited to lagging chromosomes in telophase RPE-1 cells. Cells were labelled to detect the indicated proteins synchronized from nocodazole release as in Fig. 1a. Graph shows percentage of lagging chromosomes that were positive for the indicated proteins in early telophase cell (when the main chromosome mass is positive for ESCRT III, PN+) and late telophase cell (when ESCRT III is no longer detected on the main chromosome mass, PN–) (n = 44, 78, 45, 31, 67, 37, each category, from two experiments). f, Near-continuous assembly of core membrane protein around the lagging chromosome. Images of 3D-SIM (left) and Imaris surface renderings (right) of an RPE-1 cell expressing emerin–GFP. Enlarged images show lagging chromosomes (yellow box) and PN (white box): DNA (blue), emerin (red), NUP133 (green) (representative of 22 cells, from two technical replicates). g, CLEM showing double-membrane NE (enlarged images in middle panels, cross sections) but reduced NPCs on intact (RFP–NLS+) and newly disrupted MN (RPF–NLS–, red arrows). An RPE-1 cell was fixed <20 min after spontaneous disruption of an MN. Left, top, DIC and fluorescence images immediately after loss of RFP–NLS signal in one of the two MN in the cell. Right, top, 70-nm thin section selected from the full series of the cell. Double-membrane NE is present on the PN as well as on the disrupted (red arrow) and intact MN. Middle row depicts NE at higher magnification. NPCs are prominent in the tangential view (bottom row) of PN (yellow arrows) but not on NLS-positive MN (blue arrows). No NPC is present on the disrupted RPF–NLS-negative MN. Representative of four cells with three intact and four newly disrupted MN, from three experiments (see Methods).
Extended Data Fig. 6 Microtubule disruption restores non-core NE assembly to lagging chromosomes only if it occurs early in anaphase.
a, Inhibition of aurora B (5 μM ZM447439) reduces spindle microtubule bundling and microtubule mass. Left, representative images of RPE-1 cells (n = 20, from two technical replicates). Right, linescan profiles of FI for α-tubulin (red) along the white dashed lines (see enlarged images of boxed regions). Scale bars, 10 µm. b, c, Parallel effects of nocodazole and aurora B inhibitor treatment (Fig. 2a) on non-core NE assembly. Live-cell/fixed cell imaging as in Fig. 2a showing that nocodazole-mediated microtubule depolymerization allows non-core NE assembly only if it occurs early in anaphase. Labelling to detect α-tubulin and LBR is in b; labelling to detect α-tubulin and NUP133 is in c. Left (b, c), representative images of RPE-1 cells exposed to DMSO or nocodazole at the indicated times after anaphase onset (cells were fixed and labelled 12 min after DMSO or nocodazole addition). Yellow arrows (b, c) indicate lagging chromosomes; white arrowhead (c) shows that even on a lagging chromosome where NUP133 (green) has largely been restored after nocodazole treatment, the region of this chromosome that remains in contact with residual microtubules (red) is depleted for NUP133. Right (b, c), experimental scheme (top; Extended Data Fig. 4e) and quantification (bottom). 12+ timepoint on x-axes includes cells that were exposed to nocodazole 12–16 min after anaphase onset. b, MN/PN FI ratio for LBR (mean with 95% CI, n = 64, 62, 54, 44, 63, 69 each time point, from three experiments). c, Fraction of lagging chromosome circumference with NUP133 (mean with 95% CI, n = 33, 29, 26, 25, 28, 68 each time point, from two experiments). ****P < 0.0001, two-tailed Mann–Whitney test. Scale bars, 10 µm. d, Formation of non-core gap is independent of the normal spatial distribution of aurora B. A non-core protein (NUP133) is excluded from the central spindle region (white arrowheads) on the chromosome mass in HeLa K cells expressing aurora B–GFP (white, in merged images; representative of 30 cells, from two technical replicates). Synchronization as in Extended Data Fig. 1e. NUP133 assembly occurred normally on the periphery of the main chromosome mass, away from the spindle, whether aurora B localized to the central spindle or was forced to remain near the chromosome mass (MKLP2 RNAi)30. Scale bars, 10 µm.
Extended Data Fig. 7 KIF4A knockdown partially restores non-core NE assembly to lagging chromosomes even when they have maximal (metaphase) levels of H3S10 phosphorylation.
a, As reported26, KIF4A knockdown largely preserves the redistribution of aurora B from centromeres to the central spindle (arrowheads) early in anaphase. Synchronization as in Fig. 1a. Left: Images from time-lapse series are shown from control (representative of 29 cells, from two experiments) or KIF4A-depleted (representative of 27 cells, from three experiments) HeLa K cells expressing aurora B–GFP (green) and RFP–H2B (red). t = 0 min, anaphase onset. Right, western blots showing depletion of KIF4A protein by siRNA in RPE-1 (top, three experiments) and HeLa K (bottom, three experiments) cells (also related to Fig. 3b, c; for gel source data see Supplementary Fig. 1). Scale bar, 10 µm. b, Restoration of LBR to some lagging chromosomes after KIF4A depletion (RPE-1 cells). Synchronization as in Fig. 1a. Left, representative images: DNA (blue), α-tubulin (red) and LBR (green). Right, MN/PN LBR FI ratio (mean with 95% CI, n = 105, 119, from three experiments). ****P < 0.0001, two-tailed Mann–Whitney test. Scale bars, 10 µm. The restoration of LBR is often continuous around MN with partially restored MN/PN FI ratio (see also Fig. 2a after aurora B inhibition), whereas the NPC assembly is commonly restored discontinuously after KIF4A depletion (Fig. 3b, c) or aurora B inhibition (Extended Data Fig. 8d). This is consistent with different localization patterns of LBR and NPC in the core domain on the main chromosome mass (Extended Data Fig. 1b, c, e), which may arise from their different mobilities within the NE. c, Live-cell imaging confirming the partial restoration of an NPC protein (NUP107) to lagging chromosomes in HeLa K cells expressing GFP–NUP107 (green) and RFP–H2B (red) after KIF4A knockdown. Synchronization as in Fig. 1a. Images from a confocal time-lapse series; arrowheads, GFP–NUP107 protein recruited to lagging chromosomes (representative of six cells, from three experiments). Time shown in min. Scale bar, 10 µm. d, Unlike KIF4A knockdown, disruption of aurora B spindle midzone localization by MKPL2 knockdown fails to restore the recruitment of NUP133 to lagging chromosomes. Synchronization as in Fig. 1a. Graph shows quantification (mean with 95% CI, n = 131, 136, 85, from two experiments). ****P < 0.0001; NS, P = 0.0616, two-tailed Mann–Whitney test. e, H3S10 phosphorylation does not block NPC and non-core NE assembly. Left, representative images of KIF4A-depleted HeLa K cells showing restoration of non-core (NUP107) assembly onto lagging chromosomes with high level of H3S10 phosphorylation (red arrowhead, enlarged images of the blue boxed region). The level of phospho-H3S10 is similar between the lagging chromosomes with (red arrowhead) or without (white arrowhead) NUP107 recruitment. To illustrate the relative difference in pH3S10 levels between PN and MN, pH3S10 has been scaled differently in the pH3S10 channel and the merged channel. Cells were synchronized as in Extended Data Fig. 1e. Scale bar, 10 µm. Middle, linescan profile of indicated proteins along the dashed line in the merged image. Right, FI of phospho-H3S10 on MN and the corresponding PN (mean with 95% CI, n = 24, 25, 25, 25, 24, from two technical replicates). In MN that have recruited NUP107 (MN/PN FI ratio >0.4), the H3S10 phosphorylation level is comparable to metaphase chromosomes. ***P = 0.0009, ****P < 0.0001; NS, P = 0.0709, P = 0.6699, left to right; two-tailed Mann–Whitney test.
Extended Data Fig. 8 Partitioning of the NE into core and non-core subdomains depends on the local organization of spindle microtubules.
a–c, 3D-SIM showing exclusion of non-core NE assembly from the region of the lagging chromosome adjacent to CHIMP4B-marked microtubules. a, Summary of SIM results after KIF4A knockdown. The ESCRT-III complex is recruited to small membrane holes at the sites where spindle microtubules intersect the reforming NE, where it is thought to be required for normal NE sealing23. We used ESCRT-III (CHMP4B) labelling to identify the NE region on the lagging chromosome that is intersected by microtubules for the SIM experiments. b, Imaris surface 3D renderings from SIM images (from c) showing recruitment of NUP133 (white) to the region of a lagging chromosome depleted of CHMP4B (green) and microtubules (red) from a KIF4A-depleted HeLa K cell (representative of four lagging chromosomes, from two experiments). c, SIM images showing that, similar to main chromosome masses during normal NE assembly23, NPCs (NUP133) are recruited to regions of the lagging chromosome lacking CHMP4B-decorated microtubules. Left, cartoon of entire lagging chromosome visualized from a KIF4A-depleted anaphase/telophase cell (microtubules in red, DNA in blue, z focal planes as indicated). Right, serial sections from a SIM z-stack of an anaphase/telophase HeLa K cell expressing CHMP4B–GFP (green) labelled for microtubules (red), NUP133 (white) and DNA (blue) after KIF4A depletion. These results are consistent with the idea that microtubules inhibit non-core NE assembly. d, KIF4A knockdown (Fig. 3b, c, Extended Data Fig. 7b–d) and early anaphase inhibition of aurora B have a similar effect on NPC and non-core NE assembly to lagging chromosomes. Experiments performed as in Fig. 2a in RPE-1 cells. Micrographs: left column, representative timelapse images of cells upon addition of ZM447439 or DMSO (live, 0 or 6 min after anaphase onset); right five columns, representative images of the same cells after fixation and labelling (12 min after treatment). For the cell exposed to ZM447439 at anaphase onset (middle row), one of the two lagging chromosomes exhibits small-scale separation of core (white arrowhead, emerin) and non-core (white arrow, NUP133) NE domains. Scale bar, 10 µm. Right, quantification (mean with 95% CI, n = 64, 72, 43, 72, 34, from two experiments). ****P < 0.0001; NS, P = 0.6932, two-tailed Mann–Whitney test.
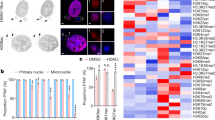
Extended Data Fig. 9 Characterization of peripherally localized missegregated chromosomes and the resulting MN: microtubule proximity, nuclear import, and chromosome number.
a, Restored assembly of NUP133 on MN derived from peripheral chromosomes as compared to central spindle lagging chromosomes in HeLa K cells. Left, quantification of the fraction of the lagging chromosome circumference with NUP133 (mean with 95% CI, n = 32, 40, from two experiments). ****P < 0.0001, two-tailed Mann–Whitney test. Right, western blots showing depletion of TTL by siRNA in HeLa K cells (three experiments; for gel source data see Supplementary Fig. 1). b, Fewer microtubules associated with peripheral missegregated chromosomes (red arrow) than with MN from central missegregated chromosomes (yellow arrow). Synchronization as in Fig. 4a. Representative images of RPE-1 cells after glutaraldehyde fixation (representative of 30 central or peripheral missegregated chromosome, from two technical replicates). Scale bars, 10 µm. The sparse microtubule density near the peripheral chromosome (bottom images) was never associated with more than three CHMP4B foci in 30 images. c–e, Normal nuclear import and DNA replication in MN from peripherally missegregated chromosomes (red arrows) as compared to MN from central missegregated chromosomes (yellow arrows). c, Top, scheme of experiment. RPE-1 cells were treated with p53 siRNA to facilitate cell-cycle progression. After mitotic exit, micronucleated cells were imaged at 20-min intervals. Bottom, images of fixed and labelled RPE-1 cells 4 h after RO release at the end of the live-cell experiment. Scale bars, 10 µm. See Fig. 4d for quantification. d, MN/PN FI ratio for lamin B1 in RPE-1 cells, 4 h after RO release (mean with 95% CI, n = 60, 32, from two experiments). ****P < 0.0001, two-tailed Mann–Whitney test. e, Restoration of DNA replication (EdU incorporation, shown as the MN/PN FI ratio) in MN from peripheral chromosomes in RPE-1 (left) and HeLa K (right) cells, 16–18 h after RO release (mean with 95% CI, n = 81, 60, from five experiments for RPE-1; n = 55, 86, from two experiments for HeLa K). In RPE-1 cells, as reported46, high dose MPS1 inhibition causes a fraction of cells to delay cell cycle progression. Therefore, we restricted analysis to cells that progressed into the S-phase of the cell cycle as evidenced by EdU labelling of the PN. **P < 0.0067, ****P < 0.0001, two-tailed Mann–Whitney test. f, MN derived from peripheral chromosomes were larger (DAPI area, top) and more decondensed (density of DAPI FI, bottom) than MN from central chromosomes. Data are from RPE-1 (left) and HeLa K (right) cells (mean with 95% CI, n = 145, 112, from three experiments for DAPI area for RPE-1, n = 145, 112, from three experiments for DAPI FI for RPE-1; n = 66, 21, from two experiments for DAPI area for HeLa K, n = 88, 58, from three experiments for DAPI FI for HeLa K). ****P < 0.0001, two-tailed Welch’s t-test for DAPI FI, two-tailed Mann–Whitney test for DAPI area. The extensive decondensation of MN from peripheral missegregated chromosomes may be because they initiate nuclear import slightly earlier than the assembling main daughter nuclei (Fig. 4c) and differ in surface area to volume ratio. g, h, Micronuclear chromosome number does not influence non-core NE assembly. Comparison of chromosome number in MN from central chromosomes and peripheral chromosomes. g, Confocal images of RPE-1 cells labelled for CENP-A (centromere marker, arrowheads), NUP133 and DNA. Yellow arrows, central chromosomes; red arrows, peripheral chromosomes. Synchronization as in Fig. 4a. Scale bars, 10 µm. Quantified in h. h, Top, distribution of chromosome number (CENP-A foci) in central (blue) and peripheral (red) MN. Data from RPE-1 (left) and HeLa K (right) cells. Bottom, fraction of lagging chromosome circumference with NUP133 for MN with the indicated chromosome number in RPE-1 (left) and HeLa K (right) cells (mean ± s.d., n = 40, 111, 20, 9, 3 each category for central chromosomes, n = 0, 6, 71, 0, 11 for peripheral chromosomes, from two experiments for RPE-1; mean ± s.d., n = 2, 35, 14, 2, 0 each category for central chromosomes, n = 0, 8, 6, 2, 0 for peripheral chromosomes, from two experiments for HeLa K). Although MN from peripheral chromosomes generally contain more chromosomes than MN from central chromosomes, non-core assembly is inhibited on MN from central chromosomes irrespective of their chromosome number. Furthermore, non-core NE assembly is restored to near-normal levels on MN from peripheral chromosomes, again irrespective of chromosome number.
Extended Data Fig. 10 Peripheral localization of lagging chromosomes reduces MN NE disruption and DNA damage; actin-dependent peripheral MN disruption in RPE-1 cells.
a, Peripheral localization of lagging chromosomes reduces MN disruption and DNA damage. Representative images of HeLa K cells (C-MN from central chromosomes; P-MN from peripheral chromosomes). See Fig. 4e for quantification. Scale bars, 5 µm. b, Detection of MN disruption by GFP–BAF accumulation yields similar results as in a for NE disruption detection by the loss of RFP–NLS. Representative images from a live-cell/fixed cell imaging experiment (as in Fig. 4e) from HeLa K cells expressing GFP–BAF. C-MN show NE disruption detected by hyper-accumulation of GFP–BAF (top) whereas P-MN show infrequent disruption (bottom). NE disruption of C-MN is accompanied by the acquisition of DNA damage (γH2AX, see Fig. 4e). At the end of 16–18 h live imaging, cells were fixed and stained for γH2AX and lamin B1. See Fig. 4e for quantification. Scale bars, 5 µm. c, In RPE-1 cells, P-MN undergo actin-dependent NE disruption. Top, experimental scheme. Chromosome missegregation was induced in RPE-1 cells expressing RFP–H2B and GFP–BAF (or GFP–H2B and RFP–NLS). About 1 h after mitotic exit, cells were treated with DMSO or a low dose (150 nM) of the actin assembly inhibitor latrunculin A (LatA) and imaged for 16–18 h. Bottom, percentage of P-MN or C-MN that underwent NE disruption (hyperaccumulation of GFP–BAF, from four experiments for DMSO and LatA) or that displayed DNA damage (FI of γH2AX in MN >3 s.d. of background γH2AX in PN, from six experiments for DMSO and three experiments for LatA). NS, P = 0.0948, **P = 0.0044, ****P < 0.0001, two-tailed Fisher’s exact test. For NE disruption: n = 258 for C- MN (DMSO), n = 176 for P-MN (DMSO), n = 173 for C-MN (LatA), n = 125 for P-MN (LatA); for DNA damage: n = 306 for C-MN (DMSO), n = 182 for P-MN (DMSO), n = 128 for C-MN (LatA), n = 73 for P-MN (LatA). d, P-MN develop discontinuities in the lamin B1 nuclear rim. Left, representative images of RPE-1 cells ~18 h after mitotic exit, as in c. Yellow boxes indicate C-MN (yellow arrowheads) with reduced lamin B1. Right, quantification (mean with 95% CI, n = 61, 28, 38, 18, left to right, from two experiments). ****P < 0.0001, two-tailed Welch’s t-test for DMSO, two-tailed Mann–Whitney test for LatA. Red boxes indicate P-MN, one of which displays a prominent lamin B1 rim discontinuity. Red arrowhead in the enlarged image shows an NE herniation on a P-MN. Scale bars, 10 µm. Consistent with prior work, MN from lagging chromosomes undergo spontaneous disruption independent of actin13. By contrast, transient disruption of PN, as occurs during confined cell migration47, is mediated by actomyosin contractile forces13. We confirm that MN from central lagging chromosomes undergo disruption independent from actin. However, we noted a difference between RPE-1 and HeLa K cells in the behaviour of MN from peripheral chromosomes. Although NE assembly appeared to be restored in both HeLa K and RPE-1 cells, P-MN in RPE-1 cells underwent residual disruption (there is nevertheless a statistically significant reduction of NE disruption frequency when comparing P-MN and C-MN in RPE-1 cells). We hypothesized that in highly motile RPE-1 cells, large P-MN might be more likely to undergo actin-dependent NE breakage, essentially becoming more similar to the transient NE disruption of PN13,47. The data in c confirm that P-MN undergo actin-dependent breakage. Furthermore, P-MN in RPE-1 cells have lamin B1 gaps (d, red arrowhead)6. One mechanism that could generate these gaps is residual contact between peripheral missegregated chromosomes and astral microtubules (Extended Data Fig. 9b). In addition, P-MN are more decondensed, and have larger NE surface area, than PN, which may dilute lamins. The increased breakage of P-MN in RPE-1 cells relative to HeLa K cells may be due to higher contractile forces in RPE-1 cells.
Supplementary information
Supplementary Figures
This file contains Supplementary Figure 1: Uncropped scans with size marker indications.
Video 1: Example of a newly formed micronucleus with normal accumulation of the nuclear import reporter, RFP-NLS (Fig. 1c, category A, representative of 5 cells from 11 experiments).
Shown are maximum intensity projections from confocal live-cell imaging of an RPE-1 cell expressing GFP-H2B and RFP-NLS. Micronuclei generation and cell synchronization were as in Fig. 1a. Images were acquired at 1-min intervals using a ×60 objective. Box indicates the micronucleus. A heat map of RFP-NLS intensity is shown in the bottom right panel.
Video 2: Example of two newly formed micronuclei with strongly defective accumulation of the nuclear import reporter, RFP-NLS (Fig. 1c, category B, representative of 9 cells from 11 experiments).
Imaging performed and analysed as in Video 1.
Video 3: Similar kinetics of CHMP4B- GFP (ESCRT-III) association and dissociation to lagging chromosome as to the main chromosome mass.
Shown are maximum intensity projections from confocal live-cell imaging of a HeLa K cell expressing CHMP4B-GFP and RFP-H2B (Extended Data Fig. 5d, representative of 10 cells from 6 experiments). Micronuclei generation and synchronization were as in Fig. 1a. Images were acquired at 1-min intervals using a ×60 objective.
Video 4: Non-core NE (GFP-NUP107) assembly on peripherally located chromosomes but not on missegregated chromosomes within the central spindle region.
Shown are maximum intensity projections from confocal live-cell imaging of a HeLa K cell expressing GFP-NUP107 and RFP-H2B. Micronuclei generation and cell synchronization were as in Fig. 4a. Images were acquired at 2-min intervals using a ×60 objective. Video corresponds to the kymograph shown in Fig. 4c [Representative of 15 cells from 3 experiments, white arrowheads, central micronuclei (C-MN); yellow arrowheads, peripheral micronuclei (P-MN)].
Video 5: Micronuclei from central chromosomes undergo spontaneous NE disruption whereas disruption is suppressed in those from peripheral chromosomes.
Shown are maximum intensity projections from wide-field live-cell imaging of an RPE-1 cell expressing GFP-BAF and RFP-H2B (4 experiments, see Extended Data Fig. 10c for quantitation). Micronuclei generation and synchronization were as in Extended Data Fig. 9c. Images were acquired at 4-min intervals for 1.5 h spanning mitosis and early interphase. Subsequently, images were acquired at 20-min intervals over 18 h. The box indicates a cell containing two micronuclei derived from central chromosomes (light blue arrows) and two micronuclei from peripheral chromosomes (red arrows). NE disruption of C-MN is evident from the sudden BAF accumulation during interphase (shown in red, ~ 18 h). Time is shown at the lower left corner.
Source data
Rights and permissions
About this article
Cite this article
Liu, S., Kwon, M., Mannino, M. et al. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 561, 551–555 (2018). https://doi.org/10.1038/s41586-018-0534-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0534-z
Keywords
This article is cited by
-
Scrambling the genome in cancer: causes and consequences of complex chromosome rearrangements
Nature Reviews Genetics (2024)
-
The two sides of chromosomal instability: drivers and brakes in cancer
Signal Transduction and Targeted Therapy (2024)
-
Aneuploidy and complex genomic rearrangements in cancer evolution
Nature Cancer (2024)
-
Genetic determinants of micronucleus formation in vivo
Nature (2024)
-
Profilin 1 deficiency drives mitotic defects and reduces genome stability
Communications Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.