Abstract
Both designed and natural anion-conducting channelrhodopsins (dACRs and nACRs, respectively) have been widely applied in optogenetics (enabling selective inhibition of target-cell activity during animal behaviour studies), but each class exhibits performance limitations, underscoring trade-offs in channel structure-function relationships. Therefore, molecular and structural insights into dACRs and nACRs will be critical not only for understanding the fundamental mechanisms of these light-gated anion channels, but also to create next-generation optogenetic tools. Here we report crystal structures of the dACR iC++, along with spectroscopic, electrophysiological and computational analyses that provide unexpected insights into pH dependence, substrate recognition, channel gating and ion selectivity of both dACRs and nACRs. These results enabled us to create an anion-conducting channelrhodopsin integrating the key features of large photocurrent and fast kinetics alongside exclusive anion selectivity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The protein coordinates and atomic structure factors of iC++ at pH 8.5 and pH 6.5 have been deposited in the Protein Data Bank (PDB) under accession number 6CSN (pH 8.5) and 6CSO (pH 6.5), respectively. The raw diffraction images have been deposited in the SBGrid data repository (ID: 570 for pH 8.5 and 571 for pH 6.5). All other data are available from the corresponding authors upon reasonable request.
References
Deisseroth, K. & Hegemann, P. The form and function of channelrhodopsin. Science 357, eaan5544 (2017).
Kato, H. E. et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482, 369–374 (2012).
Berndt, A., Lee, S. Y., Ramakrishnan, C. & Deisseroth, K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science 344, 420–424 (2014).
Wietek, J. et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science 344, 409–412 (2014).
Wietek, J. et al. An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo. Sci. Rep. 5, 14807 (2015).
Berndt, A. et al. Structural foundations of optogenetics: Determinants of channelrhodopsin ion selectivity. Proc. Natl Acad. Sci. USA 113, 822–829 (2016).
Govorunova, E. G., Sineshchekov, O. A., Janz, R., Liu, X. & Spudich, J. L. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 (2015).
Kim, C. K., Adhikari, A. & Deisseroth, K. Integration of optogenetics with complementary methodologies in systems neuroscience. Nat. Rev. Neurosci. 18, 222–235 (2017).
Gunaydin, L. A. et al. Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392 (2010).
Yizhar, O. et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178 (2011).
Kim, Y.S. et al. Crystal structure of a natural anion-conducting channelrhodopsin, GtACR1. Nature https://doi.org/10.1038/s41586-018-0511-6 (2018).
Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 (2015).
Allen, W. E. et al. Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155 (2017).
Chung, S. et al. Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 545, 477–481 (2017).
Mamad, O. et al. Place field assembly distribution encodes preferred locations. PLoS Biol. 15, e2002365 (2017).
Selimbeyoglu, A. et al. Modulation of prefrontal cortex excitation/inhibition balance rescues social behavior in CNTNAP2-deficient mice. Sci. Transl. Med. 9, eaah6733 (2017).
Sineshchekov, O. A., Li, H., Govorunova, E. G. & Spudich, J. L. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc. Natl Acad. Sci. USA 113, E1993–E2000 (2016).
Takemoto, M. et al. Molecular dynamics of channelrhodopsin at the early stages of channel opening. PLoS One 10, e0131094 (2015).
VanGordon, M. R., Gyawali, G., Rick, S. W. & Rempe, S. B. Atomistic study of intramolecular interactions in the closed-state channelrhodopsin chimera, C1C2. Biophys. J. 112, 943–952 (2017).
Govorunova, E. G. et al. The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep. 7, 43358 (2017).
Jun, J. J. et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017).
Yamashita, K., Hirata, K. & Yamamoto, M. KAMO: towards automated data processing for microcrystals. Acta Crystallogr. D Struct. Biol. 74, 441–449 (2018).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Gradinaru, V. et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010).
Zhang, L. & Hermans, J. Hydrophilicity of cavities in proteins. Proteins 24, 433–438 (1996).
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006).
Betz, R. Dabble v.2.6.3 https://doi.org/10.5281/zenodo.836914 (2004).
Best, R. B., Mittal, J., Feig, M. & MacKerell, A. D. Jr. Inclusion of many-body effects in the additive CHARMM protein CMAP potential results in enhanced cooperativity of alpha-helix and beta-hairpin formation. Biophys. J. 103, 1045–1051 (2012).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Huang, J. & MacKerell, A. D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Zhu, S., Brown, M. F. & Feller, S. E. Retinal conformation governs pK a of protonated Schiff base in rhodopsin activation. J. Am. Chem. Soc. 135, 9391–9398 (2013).
Salomon-Ferrer, R., Gotz, A. W., Poole, D., Le Grand, S. & Walker, R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Case, D. A. et al. AMBER 2017 (University of California, San Francisco, 2017).
Hopkins, C. W., Le Grand, S., Walker, R. C. & Roitberg, A. E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015).
Roe, D. R. & Cheatham, T. E. III. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Vanommeslaeghe, K. et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010).
Steinbrecher, T., Latzer, J. & Case, D. A. Revised AMBER parameters for bioorganic phosphates. J. Chem. Theory Comput. 8, 4405–4412 (2012).
Paxinos, G., Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates 2nd edn (Academic Press, San Diego, 2001).
Zhang, F. et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–7 (2004).
Acknowledgements
We thank C. Lee, M. Lo, K. Geiselhart and M. Lima for technical support; K.K. Kumar, N.R. Latorraca, M. Inoue and K. Katayama for comments on the manuscript; E.E. Steinberg, M.A. Wright, and R.C. Malenka for inputs on mouse experiments; K. Hirata, M. Yamamoto and other staffs at BL32XU of SPring-8, and the staff at BL23ID-B/23ID-D of APS for assistance in data collection. We acknowledge support by JST PRESTO (JPMJPR1782 for H.E.K., JPMJPR15P2 for K.I.), Stanford Bio-X and the Kwanjeong Foundation (Y.S.K.), German Academic Exchange Service (D.H.); MEXT (17H03007 for K.I. and 25104009/15H02391 for H.K.), JST CREST (JPMJCR1753, H.K.), the US Department of Energy, Scientific Discovery through Advanced Computing (SciDAC) program (R.O.D.) and Mathers Charitable Foundation (B.K.K.). The project was supported by a grant for channelrhodopsin crystal structure determination from the NIMH (R01MH075957 to K.D.).
Reviewer information
Nature thanks P. Scheerer, L. Tian and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
H.E.K. and Y.S.K. contributed equally and either has the right to list himself first in bibliographic documents. H.E.K. and Y.S.K. expressed, purified, and crystallized iC++, harvested crystals and collected diffraction data. H.E.K. and K.Y. solved the structures and analysed electron densities. H.E.K. and Y.S.K. measured UV-vis spectra of iC++ mutants. Y.S.K. and L.E.F. performed electrophysiology. J.M.P. performed and analysed molecular dynamics simulations under guidance of R.O.D. W.E.A. and K.E.E. conducted in vivo recording. S.I. and K.I. measured UV-vis spectra and performed flash photolysis under guidance of H.K. C.Ra. and K.E.E. performed neuron cultures and molecular cloning. C.Ri. and Y.S.K. performed C. elegans experiments under guidance of K.S. D.H. performed crystallography and S.Y.L. and A.B. performed electrophysiology at an early stage. K.D. initiated and supervised this channelrhodopsin structure-function project. H.E.K., Y.S.K., B.K.K. and K.D. planned and guided the work, and interpreted the data. H.E.K., Y.S.K. and K.D. prepared the manuscript and wrote the paper with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Spectroscopic and structural characterization of iC++.
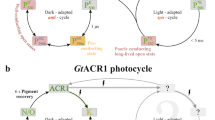
a, Sequence alignment. iC++ mutations and corresponding residues in the other opsins are highlighted. b, c, Transient absorption spectra of iC++ (b) and time traces of the absorption changes (c) at specific probe wavelengths. d, Photocycle schematic for iC++, determined from analysis of results shown in b and c. e, Overlay of the M-intermediate state measured in b (blue) and a flash-induced photocurrent generated by iC++ in a HEK293 cell (red) by flashing 100 μs, 470 nm light at 2 mW mm−2. The electrophysiology experiment was repeated independently 4 times with similar results, and the spectroscopy experiment was performed once. f, Table describing data collection and refinement statistics of iC++ in pH 6.5 and pH 8.5 conditions. Datasets were collected from 16 (pH 8.5) and 27 (pH 6.5) crystals. Values in parentheses are for the highest resolution shell.
Extended Data Fig. 2 Structural basis of pH dependence.
a, Bright-field images of iC++ crystals formed under pH 6.5 (top) and pH 8.5 (bottom) conditions. b, Overlay of iC++ crystal structures at pH 6.5 (cyan) and pH 8.5 (orange). c, Comparison of calculated mean electron density of ATR atoms (red circle) to the distribution corresponding to protein residues (box-and-whisker plots) at pH = 6.5 (cyan) or 8.5 (orange). Note lower mean electron density of ATR at pH 8.5 despite higher electron density of the overall structure. Mean values at atomic positions of the 2Fo − Fc map were calculated using phenix.get_cc_mtz_pdb in Phenix26. Box plots show median (centre), first and third interquartile ranges, minimum and maximum. Sample size (number of residues used in calculation), n = 282 for pH 8.5, and 283 for pH 6.5. Electron-density values were normalized in the model region, and maps were generated at 3.2 Å resolution. d, e, Absorption spectrum of iC++ measured over the range from pH 3.0 to pH 8.5 (d), and 483 nm and 386 nm absorbance traces collected across the measured pH range (e). Absorbance was measured for every 0.5 pH unit change while the sample was titrated by HCl. Note increased absorption at 386 nm and decreased absorption at 483 nm under alkaline conditions.
Extended Data Fig. 3 Electron densities of side chains and putative water molecules of iC++ structure at pH 8.5.
Shown are 2Fo − Fc maps (blue mesh, contoured at 1σ) for ten introduced mutations, and for two putative water molecules as labelled. All ten mutations have well-resolved electron densities to reliably position structural features, which support the interpretation of the roles of introduced mutations.
Extended Data Fig. 4 Ion-conducting pathways.
Ion-conducting pathways of C1C2 (left), GtACR1 (middle) and iC++ (right). Black meshes represent the extracellular and intracellular vestibules. Red dashed circles represent ECSs. Note that EV1 and EV2 of iC++ are structurally more similar to the extracellular vestibules of GtACR1 than those of C1C2.
Extended Data Fig. 5 Computational and crystallographic estimation of the potential chloride-binding site in iC++ at pH 8.5.
a, The r.m.s.d. of the atomic positions of α-carbons between the crystallographic structures and structures from molecular dynamics simulations was used to assess the stability of the crystal structures in a membrane environment and the quality of our molecular dynamics simulation parameters. The r.m.s.d. values computed from five independent simulations, each over 1500 ns in length, of iC++ (left) and GtACR1 (right), average less than 2 Å, supporting the stability of the crystal structures. In our calculations, we removed the flexible C-terminal tails of both proteins (residue 312 and 251 onwards in iC++ and GtACR1, respectively) and the first eight residues of GtACR1. b, 2Fo − Fc map (blue mesh, contoured at 1σ) and Fo − Fc omit map (green mesh, contoured at 4σ) for electron density around the putative chloride-binding site. c, d, 2Fo − Fc map for the same site, with a chloride ion (c) and with a phosphate (d). Note the strong negative peak observed around phosphate ion. Numbers indicate distance between two atoms connected by dashed lines. e, Chlorides frequently occupy a site near Arg281 and Arg156 on the extracellular side of iC++, during molecular dynamics simulation. The left panel depicts this binding site as a grey sphere from the perspective looking out from the dimer interface with the extracellular side at the top. Arg281 and Arg156 are shown as sticks. The other two panels show intervals of simulation in which a chloride (middle) or phosphate (right) are present in the binding site. Each horizontal bar represents an individual replicate and monomer. Time periods where a chloride or phosphate bound are coloured green and orange, respectively, and the period after the end of the simulation is coloured black.
Extended Data Fig. 6 Photocurrent densities and kinetics of wild-type and mutant ACRs.
a, b, Summary of current densities of wild-type (WT) iC++ and its mutants (a) and wild-type GtACR1 and its mutants (b) from this study. Data are mean ± s.e.m; one-way ANOVA followed by Dunnett’s test. *P < 0.05, **P < 0.01 and ***P = 0.0001. c, Photocurrent comparisons of wild-type ZipACR and N256Q mutant (left) and wild-type GtACR2 and N235Q mutant (right). Data are means ± s.e.m.; two-tailed t-test, **P = 0.0025. d, Fast (left) and slow (right) off-kinetics comparison of wild-type GtACR2 and the N235Q mutant, Data are mean ± s.e.m; two-tailed t-test, **P = 0.0014. e, Example traces of wild-type GtACR2 and the N235Q mutant expressed in HEK293 cells by lipofectamine transfection, measured at −10 mV holding potential in voltage-clamp. Traces were recorded while cells were stimulated with 1.5 s of 1.0 mW mm−2 irradiance at 470 nm for iC++ and GtACR2 and 513 nm for GtACR1 and ZipACR. Sample size (number of cells) for each experiment is indicated next to label in parentheses.
Extended Data Fig. 7 Current–voltage relationships of ACR mutants.
Current–voltage relationships from −95 mV to +15 mV were determined from light-evoked current amplitude at the indicated holding potentials. Each measurement was normalized to the current amplitude measured at −5 mV for iC++ variants and −25 mV for GtACR1 variants. Values are means and s.e.m. For iC++ variants: n = 8 for wild type, and n = 5 for the other iC++ variants. For GtACR1 variants: n = 9 for E68Q, n = 7 for R192E, n = 5 for K24A, K33A, K33E, R53Q, K56A, R83Q, K116A, K188A, K188E, R192A, K256A, Q46A/K188A, K188A/R259A, and n = 4 for the other variants. HEK293 cells expressing proteins through lipofectamine transfection method were recorded while stimulated by 1.5 s of 1.0 mW/mm2 irradiance at 470nm for iC++ and 513 nm for GtACR1. The first five graphs are from the iC++ backbone, and the rest are GtACR1 mutations.
Extended Data Fig. 8 Representative traces for current–voltage measurements.
Voltage-clamp traces corresponding to current–voltage relationships shown in Extended Data Fig. 7, collected from −95 mV to +15 mV in steps of 10 mV. HEK293 cells, transfected using the lipofectamine transfection method, were recorded while stimulated by 1.5 s of 1.0 mW mm−2 irradiance at 470 nm for iC++ and 513 nm for GtACR1. The first six traces are from the iC++ backbone, and the rest are GtACR1 mutations.
Extended Data Fig. 9 Electrophysiology and structural comparison of FLASH and ZipACR.
a, Confocal images of FLASH and ZipACR expression in mouse hippocampus 4 weeks after virus injection. Up to 15 slices per animals were collected, all with similar expression level of opsins. Scale bar, 100 μm. b–h, Comparisons of input resistance (b), capacitance (c), resting potentials (d), photocurrent measured at −10 mV (e), reversal potentials (f), currents injected to elicit action potentials (g), and holding currents needed to keep cells at −70 mV (h) The steady-state/peak photocurrent ratio of ZipACR was close to 100%, so the stationary current data was not included in e. All electrophysiology data were measured from FLASH- and ZipACR-expressing hippocampal neurons in acute slice preparation from animals 4 weeks after virus injection. Data are mean ± s.e.m.; two-tailed t-test. *P = 0.044, **P = 0.003. i, Example traces of neurons expressing FLASH and ZipACR succeeding (left) and failing (right) to inhibit multiple spikes under the pulsed illumination condition. j, Summary of FLASH and ZipACR multiple-spike inhibition, corresponding to the experimental paradigm in i. Data are mean ± s.e.m.; two-tailed t-test. P = 0.32. k, Light-driven spiking was observed from ZipACR expressing neurons (3/8), but not from FLASH expressing ones (0/8). For all electrophysiology experiments, sample size (number of cells) for each experiment is indicated next to label in parentheses. l, m, Surface electrostatic potentials of homology models of FLASH (l) and ZipACR (m). Models were built using SWISS-MODEL47. The surface is coloured on the basis of electrostatic potential contoured from −10 kT (red) to +10 kT (blue). White denotes 0 kT. Surface potential was calculated using PDB2PQR48 for both GtACR1 and C1C2 models.
Extended Data Fig. 10 Confocal images of opsin expression in brain slices and worms.
a, b, Confocal images showing opsin expression with electrode (a) and in thalamus (b). Scale bar, 100 μm. c, d, Confocal images showing FLASH (c) and ZipACR (d) expression in worms, mediated by punc-17- (cholinergic neurons, top) and pmyo-3- (muscle, bottom) promoters. Scale bar, 20 μm.
Extended Data Fig. 11 Differences in polarity and hydration of retinal binding pockets in iC++ and GtACR1.
a, Structures of the retinal binding pockets of GtACR1 (left) and iC++ (right) in van der Waals representations. b, Absorption spectra of wild-type iC++, S295A, G220S and S295A/G220S mutants. Water density map at the dimer interface of GtACR1, viewed parallel to and from within the membrane (c) and viewed from the intracellular side (d) during molecular dynamics simulation, contoured at probability density of 0.016 molecules per Å3. e, Corresponding water density map at the dimer interface for iC++ viewed parallel to and from within the membrane, contoured at probability density of 0.016 molecules per Å3. f, Superimposed structures of iC++ (beige) and GtACR1 (blue) at the dimer interfaces, viewed from the intracellular side. ATR molecules are shown as sphere models.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion.
Source data
Rights and permissions
About this article
Cite this article
Kato, H.E., Kim, Y.S., Paggi, J.M. et al. Structural mechanisms of selectivity and gating in anion channelrhodopsins. Nature 561, 349–354 (2018). https://doi.org/10.1038/s41586-018-0504-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0504-5
Keywords
This article is cited by
-
A blue-shifted anion channelrhodopsin from the Colpodellida alga Vitrella brassicaformis
Scientific Reports (2023)
-
Aion is a bistable anion-conducting channelrhodopsin that provides temporally extended and reversible neuronal silencing
Communications Biology (2022)
-
Preference of Proteomonas sulcata anion channelrhodopsin for NO3− revealed using a pH electrode method
Scientific Reports (2021)
-
Small-wedge synchrotron and serial XFEL datasets for Cysteinyl leukotriene GPCRs
Scientific Data (2020)
-
Nano-enabled cellular engineering for bioelectric studies
Nano Research (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.