Abstract
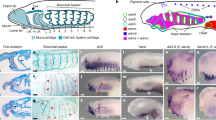
Placodes and neural crests represent defining features of vertebrates, yet their relationship remains unclear despite extensive investigation1,2,3. Here we use a combination of lineage tracing, gene disruption and single-cell RNA-sequencing assays to explore the properties of the lateral plate ectoderm of the proto-vertebrate, Ciona intestinalis. There are notable parallels between the patterning of the lateral plate in Ciona and the compartmentalization of the neural plate ectoderm in vertebrates4. Both systems exhibit sequential patterns of Six1/2, Pax3/7 and Msxb expression that depend on a network of interlocking regulatory interactions4. In Ciona, this compartmentalization network produces distinct but related types of sensory cells that share similarities with derivatives of both cranial placodes and the neural crest in vertebrates. Simple genetic disruptions result in the conversion of one sensory cell type into another. We focused on bipolar tail neurons, because they arise from the tail regions of the lateral plate and possess properties of the dorsal root ganglia, a derivative of the neural crest in vertebrates5. Notably, bipolar tail neurons were readily transformed into palp sensory cells, a proto-placodal sensory cell type that arises from the anterior-most regions of the lateral plate in the Ciona tadpole6. Proof of transformation was confirmed by whole-embryo single-cell RNA-sequencing assays. These findings suggest that compartmentalization of the lateral plate ectoderm preceded the advent of vertebrates, and served as a common source for the evolution of both cranial placodes and neural crest3,4.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Northcutt, R. G. & Gans, C. The genesis of neural crest and epidermal placodes: a reinterpretation of vertebrate origins. Q. Rev. Biol. 58, 1–28 (1983).
Baker, C. V. & Bronner-Fraser, M. The origins of the neural crest. Part II: an evolutionary perspective. Mech. Dev. 69, 13–29 (1997).
Schlosser, G. Do vertebrate neural crest and cranial placodes have a common evolutionary origin? BioEssays 30, 659–672 (2008).
Schlosser, G., Patthey, C. & Shimeld, S. M. The evolutionary history of vertebrate cranial placodes II. Evolution of ectodermal patterning. Dev. Biol. 389, 98–119 (2014).
Stolfi, A., Ryan, K., Meinertzhagen, I. A. & Christiaen, L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 527, 371–374 (2015).
Wagner, E., Stolfi, A., Gi Choi, Y. & Levine, M. Islet is a key determinant of ascidian palp morphogenesis. Development 141, 3084–3092 (2014).
Manni, L. et al. Neurogenic and non-neurogenic placodes in ascidians. J. Exp. Zool. B Mol. Dev. Evol. 302B, 483–504 (2004).
Mazet, F. et al. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev. Biol. 282, 494–508 (2005).
Pasini, A. et al. Formation of the ascidian epidermal sensory neurons: insights into the origin of the chordate peripheral nervous system. PLoS Biol. 4, e225 (2006).
Imai, J. H. & Meinertzhagen, I. A. Neurons of the ascidian larval nervous system in Ciona intestinalis: II. Peripheral nervous system. J. Comp. Neurol. 501, 335–352 (2007).
Horie, T., Kusakabe, T. & Tsuda, M. Glutamatergic networks in the Ciona intestinalis larva. J. Comp. Neurol. 508, 249–263 (2008).
Wagner, E. & Levine, M. FGF signaling establishes the anterior border of the Ciona neural tube. Development 139, 2351–2359 (2012).
Patthey, C., Schlosser, G. & Shimeld, S. M. The evolutionary history of vertebrate cranial placodes—I: cell type evolution. Dev. Biol. 389, 82–97 (2014).
Abitua, P. B. et al. The pre-vertebrate origins of neurogenic placodes. Nature 524, 462–465 (2015).
Imai, K. S., Levine, M., Satoh, N. & Satou, Y. Regulatory blueprint for a chordate embryo. Science 312, 1183–1187 (2006).
Tresser, J. et al. doublesex/mab3 related-1 (dmrt1) is essential for development of anterior neural plate derivatives in Ciona. Development 137, 2197–2203 (2010).
Aniello, F. et al. Identification and developmental expression of Ci-msxb: a novel homologue of Drosophila msh gene in Ciona intestinalis. Mech. Dev. 88, 123–126 (1999).
Wada, H., Holland, P. W., Sato, S., Yamamoto, H. & Satoh, N. Neural tube is partially dorsalized by overexpression of HrPax-37: the ascidian homologue of Pax-3 and Pax-7. Dev. Biol. 187, 240–252 (1997).
Huang, X., Hong, C. S., O’Donnell, M. & Saint-Jeannet, J. P. The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl Acad. Sci. USA 102, 11349–11354 (2005).
Parlier, D. et al. The Xenopus doublesex-related gene Dmrt5 is required for olfactory placode neurogenesis. Dev. Biol. 373, 39–52 (2013).
Ahrens, K. & Schlosser, G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev. Biol. 288, 40–59 (2005).
Schlosser, G. & Ahrens, K. Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 271, 439–466 (2004).
Pieper, M., Eagleson, G. W., Wosniok, W. & Schlosser, G. Origin and segregation of cranial placodes in Xenopus laevis. Dev. Biol. 360, 257–275 (2011).
Köster, M., Dillinger, K. & Knöchel, W. Activin A signaling directly activates Xenopus winged helix factors XFD-4/4′, the orthologues to mammalian MFH-1. Dev. Genes Evol. 210, 320–324 (2000).
Bailey, A. P., Bhattacharyya, S., Bronner-Fraser, M. & Streit, A. Lens specification is the ground state of all sensory placodes, from which FGF promotes olfactory identity. Dev. Cell 11, 505–517 (2006).
Berry, F. B. et al. Functional interactions between FOXC1 and PITX2 underlie the sensitivity to FOXC1 gene dose in Axenfeld–Rieger syndrome and anterior segment dysgenesis. Hum. Mol. Genet. 15, 905–919 (2006).
Corbo, J. C., Levine, M. & Zeller, R. W. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589–602 (1997).
Schlosser, G. Induction and specification of cranial placodes. Dev. Biol. 294, 303–351 (2006).
Abitua, P. B., Wagner, E., Navarrete, I. A. & Levine, M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 492, 104–107 (2012).
Gainous, T. B., Wagner, E. & Levine, M. Diverse ETS transcription factors mediate FGF signaling in the Ciona anterior neural plate. Dev. Biol. 399, 218–225 (2015).
Russo, M. T. et al. Regulatory elements controlling Ci-msxb tissue-specific expression during Ciona intestinalis embryonic development. Dev. Biol. 267, 517–528 (2004).
Roure, A., Lemaire, P. & Darras, S. An otx/nodal regulatory signature for posterior neural development in ascidians. PLoS Genet. 10, e1004548 (2014).
Hozumi, A. et al. Enhancer activity sensitive to the orientation of the gene it regulates in the chordate genome. Dev. Biol. 375, 79–91 (2013).
Satou, Y., Imai, K. S. & Satoh, N. Action of morpholinos in Ciona embryos. Genesis 30, 103–106 (2001).
Satou, Y., Imai, K. S. & Satoh, N. The ascidian Mesp gene specifies heart precursor cells. Development 131, 2533–2541 (2004).
Horie, T. et al. Ependymal cells of chordate larvae are stem-like cells that form the adult nervous system. Nature 469, 525–528 (2011).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Satou, Y., Kawashima, T., Shoguchi, E., Nakayama, A. & Satoh, N. An integrated database of the ascidian, Ciona intestinalis: towards functional genomics. Zool. Sci. 22, 837–843 (2005).
Acknowledgements
We thank all members of the LSI genome facility for technical support of the single-cell RNA-seq assays and analysis; R. Yoshida and C. Imaizumi and all other members of staff at the Maizuru Fisheries Research Station of Kyoto University for providing Ciona intestinalis. This study was supported by a grant from the NIH to M.L. (NS076542) and by Grants-in-Aid for Scientific Research from JSPS to T.H. (24687008, 16K07433). T.H. was supported by the Pre-Strategic Initiatives, University of Tsukuba. A portion of Ciona intestinalis and plasmids used in this study were provided by the National Bio-Resource Project (NBRP) of the MEXT, Japan. A.H. was partially supported by a fellowship from the Colombian Government (Colciencias 568).
Reviewer information
Nature thanks N. Satoh, G. Schlosser, S. Shimeld and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
T.H., A.H. and M.L. conceived the project. T.H., K.C. and M.L. designed the experiments. R.H., K.C. and T.H. performed the experiments. R.H., A.H., K.C., C.C., T.H. and M.L. analysed and interpreted the data. T.H., A.H., K.C., C.C. and M.L. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sensory cell lineages.
a, Cell lineages of anterior blastomeres (a-line blastomeres) and a posterior blastomere (b-line) from the 16-cell to 110-cell stages. Green, Dmrt.a-expressing blastomeres; magenta, Msxb expression lineages; yellow, Foxc expression. b, Schematic of 16-cell-stage embryos. Each of the blastomeres that was labelled with Dil or DiO is indicated by magenta or green, respectively. c–e, Head regions of larvae labelled with DiI or DiO at the 16-cell stage. c, Labelling of the a5.3 lineage. d, Labelling of the a5.4 lineage. e, Labelling of the b5.3 blastomere. f–h, Head region of a larva that was injected with Dmrt.a > CFP and Msxb > mCherry reporter genes. Arrowhead identifies the boundary of the Dmrt.a–Msxb expression territories. i–k, Head region of a larva injected with Foxc > CFP and Dmrt.a > mCherry reporter genes. Anterior is to the left. Scale bars, 100 μm.
Extended Data Fig. 2 Regulation of Eya expression by Dmrt.a and Msxb.
a–c, Head regions of larvae injected with an Eya > CFP reporter gene. Yellow arrowheads denote Eya expression in the proto-placodal region of control MO-injected larvae (36 out of 36 larvae displayed this pattern) (a). b, There is a loss of expression in Dmrt.a morphants (88 out of 88 larvae showed this phenotype). c, There is expanded expression (white arrowheads) in Msxb morphants (39 out of 48 larvae showed this phenotype). Anterior is to the left. Scale bars, 100μm.
Extended Data Fig. 3 Regulatory interactions among placodal determinants.
a, b, Head regions of larvae that were injected with an Otx MO, and injected with Six1/2 > mCherry (a) and Eya > CFP (b) (32 out of 32 larvae showed reduced or no expression of Six1/2 > mCherry and no expression of Eya > CFP). c, d, Head regions of larvae injected with Dmrt.a > Msxb, and injected with Six1/2 > CFP (c; 33 out of 33 larvae showed no expression of Six1/2 > CFP) and Eya > CFP (d; 71 out of 71 larvae showed little or no expression of Eya > CFP). e, f, Head regions of larvae injected with Six1/2 > mCherry, and injected with H2O (e; 61 out of 61 larvae showed full expression of Six1/2 > mCherry) or Dmrt.a > Foxc (f; 50 out of 50 larvae showed no expression of Six1/2 > mCherry). g, h, Head regions of larvae injected with Foxc > mCherry, and injected with H2O (g; 40 out of 40 larvae showed full expression of Foxc > mCherry) or Dmrt.a > Six1/2 (h; 39 out of 39 larvae showed no expression of Foxc > mCherry). Scale bars, 100 μm.
Extended Data Fig. 4 Direct repression of Six1/2 expression by Msxb.
a, Deletion analyses of the 5′ regulatory region of Six1/2. −2,410 bp to −2,001 bp of the 5′ cis-regulatory region is necessary for Kaede expression in the pre-placodal territory. b, c, Head regions of larvae injected with Six1/2 −2,410 to −2,001 > Kaede, and injected with H2O (b; 74 out of 74 larvae showed full expression of Six1/2 −2,410 to −2,001 > Kaede) or Dmrt.a > Msxb (c; 37 out of 37 larvae showed no expression of Six1/2 −2,410 to −2,001 > Kaede). d, The Six1/2 5′ regulatory region spanning −2,410 to −2,001 bp contains an Otx binding site (green box) and multiple Msxb repressor binding sites (magenta boxes).
Extended Data Fig. 5 Anterior expansion of Msxb expression in Dmrt.a morphant.
Tailbud embryo injected with Dmrt.a MO, Dmrt.a ΔMO target sequence > CFP and Msxb > mCherry construct. The anterior expansion of the Msxb > mCherry expression pattern is indicated by the white arrowhead. Anterior is to the left. Scale bars, 100 μm.
Extended Data Fig. 6 Specification of aATEN sensory neurons.
a–c, Larvae injected with the CNGA > CFP reporter gene. Yellow arrowheads indicate expression in aATENs, and white arrowheads indicate ectopic sites of differentiated aATENs in the palp. a, Larva injected with Dmrt.a MO (62 out of 62 larvae showed no expression of CNGA > CFP in aATENs). b, Larva injected with control MO (32 out of 32 larvae showed full expression of CNGA > CFP). c, Larva injected with Foxc MO (18 out of 42 larvae showed expanded expression of CNGA > CFP in palp region). Anterior is to the left. Scale bars, 100 μm.
Extended Data Fig. 7 Single-cell RNA-seq analysis.
a, b, Identification of major cell types among 20 clusters with known markers, 3 epidermis clusters were identified by the expression of EpiB; 2 clusters of epidermis and sensory neurons were identified by the expression of either Msxb or nut, along with EpiB; a cluster of sensory neurons identified by the expression of NGN3 and nut; 3 clusters of central nervous system by high levels of nut transcripts; 6 clusters of mesenchyme by their expression of twist; 2 endoderm clusters by the high expression of SOD3; and one notochord cluster by the expression of CiT (T also known as brachyury). c, Representation of overlap between cells from Pax3/7 > Foxc and control embryos in each cell population. t-SNE plot from Fig. 3a, with each cell now coloured to indicate their origin from either Pax3/7 > Foxc embryos (red dots, n = 5,339) or control embryos (blue dots, n = 4,850). Both samples contribute to all 20 cell populations. d, Identification of BTNs and PSCs with the combination of representation markers in the control embryo, 27 BTNs (green dots) were identified by the combination of Asic1b and synaphin, and 15 PSCs were identified by the combination of Foxg, islet and SP8. e, Visualization of SV40+ cells in Pax3/7 > Foxc transgenic embryos within t-SNE projection map. SV40 is detected in cells contained within clusters 2 and 5 for epidermis and sensory neuron cells, as well as weak expression in mesenchyme clusters 15 and 16. None of the transformed or hybrid cells contained in the sensory cell clusters (5 and 6) express any mesenchyme marker genes, suggesting that none of these are transformed by misexpression of Pax3/7. f, Heat map of representative genes (Fig. 3e) that show no significant differential expression in SV40+ cells contained within clusters 2, 5, 15 and 16. g, Heat map of all differentially expressed genes between BTNs and PSCs from both control and Pax3/7 > Foxc embryos.
Extended Data Fig. 8 Newly identified markers for PSCs and BTNs.
Distribution of newly identified marker genes in PSCs (a) and BTNs (b).
Extended Data Fig. 9 Heat map of differentially expressed and coexpressed genes between PSCs, aATENs and BTNs from wild-type late tailbud stage II embryos.
Transcription factors (red), signalling pathway genes (green) and effector genes (black).
Extended Data Fig. 10 Control experiments for MO gene disruption assays.
a–d, Dmrt.a MO. a, Schematic of reporter gene containing Dmrt.a regulatory genes with and without recognition sequences for the Dmrt.a MO that was used in this study. The MO recognition sequences are located in the 5′ UTR, upstream of the initiating AUG codon (−1). b, Larva injected with Dmrt.a MO and injected with the Dmrt.a > CFP reporter gene containing the MO recognition sequences (46 out of 46 larvae showed no protein synthesis from the Dmrt.a > CFP reporter). c, Same as b, except that the reporter gene lacks the Dmrt.a MO recognition sequence (Dmrt.a ΔMO target > CFP) (57 out of 57 larvae showed CFP expression in appropriate head tissues). Dmrt.a MO efficiently blocks the expression of CFP that contains the Dmrt.a MO target sequence. d, Larva injected with Dmrt.a MO, Dmrt.a ΔMO > Dmrt.a and Six1/2 > mCherry. Dmrt.a MO morphants normally lack Six1/2 > mCherry expression (Fig. 2b), but expression is restored with a Dmrt.a transgene that lacks the MO recognition sequence (107 out of 108 larvae showed expression of Six1/2 > mCherry). This result shows that the Dmrt.a MO used in this study specifically blocks the synthesis of Dmrt.a protein products. e–h, Msxb MO. e, Diagram of Msxb 5′ regulatory region and the location of the recognition sequences for the Msxb MO and point mutations in this sequence. f, g, Larvae injected with Msxb > CFP containing MO recognition sequences, and injected with control MO (f; 54 out of 54 larvae showed CFP expression). g, Same as f except that the Msxb MO was injected instead of the control MO (44 out of 44 larvae showed no expression of CFP). These results show that the Msxb MO specifically blocks CFP protein synthesis from the Msxb reporter gene. h, Larva injected with Msxb MO, Six1/2 > mCherry and Msxb > CFP reporter gene containing point mutations in MO recognition sequence (see red letters in e). Msxb morphants normally display expanded expression of Six1/2 in tail regions (Fig. 2c). This expansion is suppressed by injection of the mutant Msxb transgene lacking the MO recognition sequences (h). This result suggests that the Msxb MO inhibits synthesis of Msxb protein products. i–l, Foxc MO. i, Diagram of Foxc 5′ regulatory region showing the location of the MO recognition sequence and point mutations in this sequence. j–l, Larvae injected with Foxc > Foxc transgene and Foxc > CFP reporter gene, and also injected with control MO (j; 25 out of 25 larvae showed CFP expression). k, Same as j except that the embryo was injected with the Foxc MO instead of the control MO (k; 99 out of 99 larvae showed no expression of CFP). This result shows that the Foxc MO efficiently blocks the synthesis of CFP proteins encoded by the Foxc > CFP reporter gene containing the Foxc MO target sequence. l, Larvae injected with Foxc MO, Foxc > Foxc mut (MO-resistant Foxc cDNA) and βγ-crystallin > mCherry. Normally, Foxc morphants lack expression of the βγ-crystallin > mCherry reporter gene (Fig. 2f). However, expression is restored by injection of Foxc > Foxc transgene. This result suggests that the Foxc MO inhibits synthesis of Foxc protein products. Scale bars, 100 μm.
Supplementary information
Supplementary Table 1
List of marker genes for all cluster in Figure 3a, BTNs marker top 100 and PSC marker top 100. This table contains the marker genes for all clusters in Figure 3a (sheet1), BTNs markers top 100 (sheet2) and PSCs markers top 100.
Rights and permissions
About this article
Cite this article
Horie, R., Hazbun, A., Chen, K. et al. Shared evolutionary origin of vertebrate neural crest and cranial placodes. Nature 560, 228–232 (2018). https://doi.org/10.1038/s41586-018-0385-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0385-7
This article is cited by
-
Ascidian embryonic cells with properties of neural-crest cells and neuromesodermal progenitors of vertebrates
Nature Ecology & Evolution (2024)
-
Shaping faces: genetic and epigenetic control of craniofacial morphogenesis
Nature Reviews Genetics (2023)
-
BMP signaling is required to form the anterior neural plate border in ascidian embryos
Development Genes and Evolution (2023)
-
Highly distinct genetic programs for peripheral nervous system formation in chordates
BMC Biology (2022)
-
Hmx gene conservation identifies the origin of vertebrate cranial ganglia
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.