Abstract
53BP1 governs a specialized, context-specific branch of the classical non-homologous end joining DNA double-strand break repair pathway. Mice lacking 53bp1 (also known as Trp53bp1) are immunodeficient owing to a complete loss of immunoglobulin class-switch recombination1,2, and reduced fidelity of long-range V(D)J recombination3. The 53BP1-dependent pathway is also responsible for pathological joining events at dysfunctional telomeres4, and its unrestricted activity in Brca1-deficient cellular and tumour models causes genomic instability and oncogenesis5,6,7. Cells that lack core non-homologous end joining proteins are profoundly radiosensitive8, unlike 53BP1-deficient cells9,10, which suggests that 53BP1 and its co-factors act on specific DNA substrates. Here we show that 53BP1 cooperates with its downstream effector protein REV7 to promote non-homologous end joining during class-switch recombination, but REV7 is not required for 53BP1-dependent V(D)J recombination. We identify shieldin—a four-subunit putative single-stranded DNA-binding complex comprising REV7, c20orf196 (SHLD1), FAM35A (SHLD2) and FLJ26957 (SHLD3)—as the factor that explains this specificity. Shieldin is essential for REV7-dependent DNA end-protection and non-homologous end joining during class-switch recombination, and supports toxic non-homologous end joining in Brca1-deficient cells, yet is dispensable for REV7-dependent interstrand cross-link repair. The 53BP1 pathway therefore comprises distinct double-strand break repair activities within chromatin and single-stranded DNA compartments, which explains both the immunological differences between 53bp1- and Rev7- deficient mice and the context specificity of the pathway.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ward, I. M. et al. 53BP1 is required for class switch recombination. J. Cell Biol. 165, 459–464 (2004).
Manis, J. P. et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5, 481–487 (2004).
Difilippantonio, S. et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456, 529–533 (2008).
Dimitrova, N., Chen, Y.-C. M., Spector, D. L. & de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008).
Cao, L. et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35, 534–541 (2009).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 (2010).
Lieber, M. R., Ma, Y., Pannicke, U. & Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4, 712–720 (2003).
Chapman, J. R. et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 (2013).
Cuella-Martin, R. et al. 53BP1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol. Cell 64, 51–64 (2016).
Di Virgilio, M. et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 (2013).
Escribano-Díaz, C. et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013).
Xu, G. et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 (2015).
Hobeika, E. et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl Acad. Sci. USA 103, 13789–13794 (2006).
Wang, J. H. et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature 460, 231–236 (2009).
Wojtaszek, J. et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ. J. Biol. Chem. 287, 33836–33846 (2012).
Hara, K. et al. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J. Biol. Chem. 285, 12299–12307 (2010).
Listovsky, T. & Sale, J. E. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J. Cell Biol. 203, 87–100 (2013).
Hakim, O. et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 484, 69–74 (2012).
Tomida, J. et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 43, 1000–1011 (2015).
Bluteau, D. et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J. Clin. Invest. 126, 3580–3584 (2016).
Fattah, F. J. et al. The transcription factor TFII-I promotes DNA translesion synthesis and genomic stability. PLoS Genet. 10, e1004419 (2014).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols 10, 845–858 (2015).
Fan, J. & Pavletich, N. P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 (2012).
Noordermeer, S. M. et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature https://doi.org/10.1038/s41586-018-0340-7 (2018).
Palm, W. & de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008).
Stavnezer, J. & Schrader, C. E. IgH chain class switch recombination: mechanism and regulation. J. Immunol. 193, 5370–5378 (2014).
Hardy, R. R., Carmack, C. E., Shinton, S. A., Kemp, J. D. & Hayakawa, K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173, 1213–1225 (1991).
Ward, I. M., Minn, K., van Deursen, J. & Chen, J. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23, 2556–2563 (2003).
Adams, I. R. & McLaren, A. Identification and characterisation of mRif1: a mouse telomere-associated protein highly expressed in germ cells and embryo-derived pluripotent stem cells. Dev. Dyn. 229, 733–744 (2004).
Skarnes, W. C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011).
Khalaj, M. et al. A missense mutation in Rev7 disrupts formation of Polζ, impairing mouse development and repair of genotoxic agent-induced DNA lesions. J. Biol. Chem. 289, 3811–3824 (2014).
Kikuchi, S., Hara, K., Shimizu, T., Sato, M. & Hashimoto, H. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J. Biol. Chem. 287, 33847–33852 (2012).
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284 (2011).
Acknowledgements
We thank members of the Chapman, Green, Cornall and Rottenberg laboratories for discussions; T. Humphrey for comments on the manuscript; D. Adams for the Rev7tm1a mouse strain; S. Boulton for 53bp1−/− mice; J. Grimes and R. Nolan for assistance with protein modelling and statistics; and B. Davies, M. Barazas, B. Reina-San-Martin, B. Deplancke, L. Vasilieva, A. Nussenzweig and E. Callen-Moreau for reagents and advice. H.G. thanks K. Ghezraoui for her unwavering support and encouragement. This project was funded by Medical Research Council (MRC) Grant (MR/M009971/1) and Cancer Research UK Career Development Fellowship (C52690/A19270) awarded to J.R.C. C.A. and M.D.-L. are funded by the MRC, R.J.C. is a Principal Investigator of the MRC Human Immunology Unit. R.F. and B.M.K. are supported by the Kennedy Trust and the John Fell Fund. M.S.-C. and E.M.-F. were supported by ERASMUS+ fellowships. The Wellcome Centre for Human Genetics is supported by Wellcome grant (090532/Z/09/Z).
Author information
Authors and Affiliations
Contributions
H.G. and C.O. designed and performed the majority of the experiments, and analysed the data with assistance from K.B., C.A., M.D.-L., M.S.-C., E.F.-M. and R.J.C. J.R.B. designed and performed the KB1P-G3 experiments, and analysed the data. K.B. performed and analysed the immunization and ELISA experiments, established the mixed chimaeras and coordinated animal experiments. D.M. and C.M.G. performed and analysed all fluorescence in situ hybridization and metaphase chromosome experiments. R.F. ran and analysed the LC–MS/MS experiments with assistance from S.B. and supervision from B.M.K. This study was initiated in collaboration with S.R. J.R.C. conceived and supervised the study, performed experiments, analysed the data, prepared the figures and wrote the manuscript with editorial contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 CSR characterization in Rev7 conditional-knockout mice.
a, PCR amplicons from genomic DNA obtained by ear biopsies or purified splenic B cells (left), or flow-cytometry-sorted cells from Hardy fractions A, B and C (right) from mice of the indicated genotype. Bands of different size correspond to the Rev7tm1c allele (cond, 475 bp), Rev7+ allele (wild type, 314 bp) and Rev7tm1d allele (flox, 255 bp). Representative data; n > 3 experiments. b, Western blot analysis of REV7 protein expression in splenic B cells isolated from mice with the indicated genotype. Representative data; n = 2 experiments. For gel source data, see Supplementary Fig. 1. c, CTV-labelled purified B cells were stimulated as indicated and stained for surface IgG1 (left) or IgG2b (centre) or IgG3 (right) on day 4. Representative of n > 6 experiments.
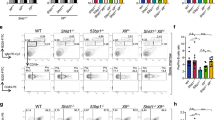
Extended Data Fig. 2 B cell lineage developmental differences in Rev7- and 53bp1-deficient mice.
a, Flow cytometry analysis of B cell development in the bone marrow of Rev7+/+Mb1+/cre, Rev7f/fMb1+/cre and 53bp1−/−Mb1+/cre mice; gating on B220+CD43+ (left, Hardy fractions A, B and C) and on B220+CD43− (right, Hardy fractions D, E and F). Representative data; n > 8 experiments. b, Apoptotic indices of total, pre-pro-B to pre-B, immature and mature B cell fractions in the bone marrow (left) and spleen (right) of Rev7+/+Mb1+/cre (n = 4), Rev7f/fMb1+/cre (n = 4), 53bp1−/−Mb1+/cre (n = 2) mice. c, Flow cytometric sub-classification of mature splenic B cell fractions in Rev7+/+Mb1+/cre (n = 8), Rev7f/fMb1+/cre (n = 8), 53bp1−/−Mb1+/cre (n = 9) mice. P values, unpaired Student’s two-tailed t-test. Bars represent mean ± 95% confidence interval. d, Top, indicated populations of pooled pro- to pre-B cell stage bone marrow lymphocytes (B220+CD43+; Hardy fraction A, B and C) from n = 2 mice per genotype were FACS-sorted and used to generate whole cell extracts. Bottom, immunoblot shows an absence of REV7 protein in extracts prepared from Rev7f/fMb1+/cre experimental bone marrow, when compared to extracts prepared from Rev7f/fMb1+/+ (no Cre) controls, yet equivalent levels of 53BP1, histone H3 (loading control) and total protein (Ponceau S stain). For gel source data, see Supplementary Fig. 1. e, Left, diagram of the mixed bone marrow chimaera transplantation experiment. Bone marrow cells from a wild-type CD45.1+ donor mouse were combined with an equal number of bone marrow cells from an experimental CD45.2+ donor mouse, and injected into lethally irradiated recipient CD45.1+ mice (n = 8 per genotype). After eight weeks, the recipient bone marrow was analysed for the relative contribution of CD45.1+ or CD45.2+ cells to reconstitute the recipient mice. Right top, enumeration of B cell precursors (as per Fig. 1g) of CD45.1+ (white circles) or CD45.2+ (from Rev7f/fMb1+/cre or 53bp1−/−Mb1+/cre mice; black circles) cells in the bone marrow after reconstitution. Right bottom, stage-specific ratios of CD45.1+ to CD45.2+ grafted B cells for indicated mixed chimaeras. In parallel, an additional control experiment involving wild-type CD45.1 and Mb1+/cre mixed chimaera was performed, resulting in equal CD45.1:CD45.2 reconstitution (data not shown). P values, multiple t-test with Holm–Sidak correction; mean ± 95% confidence interval. INP, input.
Extended Data Fig. 3 REV7 protein expression and CSR in complemented Rev7−/− CH12-F3 cells.
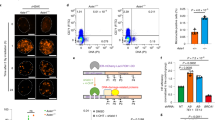
a, Stable Flag–HA–REV7 protein expression in indicated complemented CH12-F3 cell lines as determined by western blotting with HA-specific antibody. Tubulin served as loading control. Representative data; n > 3 independent experiments. For gel source data, see Supplementary Fig. 1. b, Flow-cytometric plots of CSR data in Fig. 2b. Representative data; n > 3 independent experiments. c, Chromatin prepared from indicated CH12-F3 lines 30 h after stimulation was subjected to ChIP with RPA34 or histone H3 (control) antibodies. Schematic of mouse Igh locus with positions of qPCR amplicons. ChIP recoveries were quantified against input DNA. Representative data, n = 2 independent experiments. Mean ± s.d.
Extended Data Fig. 4 Distinct REV7 residues mediate its functions in interstrand cross-link repair.
a, Indicated REV7 mutants were tested for their interaction with the RBM1 of REV3L by yeast two-hybrid assay. A fragment of REV3L (amino acids 1775–2200) with RBM2 mutated (P1996A and P2001A) was used. Fivefold dilutions, n = 4 biological repeats. b, Representative histograms of indicated CH12-F3 lines, mock- or MMC-treated for 8 and 24 h. Numbers depict sub-G1 and G2/M populations as a proportion (%) of total events. Representative data, n = 3 independent experiments. c, Representative propidium iodide and DAPI co-stained metaphases from indicated CH12-F3 cell lines after 24 h incubation with MMC; n = 2 independent experiments. Arrows, chromosome breaks (red) and radial chromosome (blue). d, Blind quantification of DSBs (chromatid and chromosome-type) and radials in indicated cell lines, with each metaphase scored as a single point. n = 2 independent experiments, each with 50 metaphases scored per genotype and condition. Bars indicate mean. e, Cell cycle of indicated CH12-F3 lines after MMC treatment. Proportions (%) of sub-G1, G1, S and G2/M events. n = 3 independent experiments ± s.d.
Extended Data Fig. 5 Identification of the REV7–shieldin complex.
a, N-terminal SHLD3 (FLJ26957) sequences contain conserved REV7-binding motifs (RBM1 and RBM2). Pairwise alignment of RBM1 of REV3L with RBM1 and RBM2 of SHLD3 across multiple species. Black and grey shading indicate identical and similar residues, respectively. b, Surface representation of REV7 crystal structure (RCSB Protein Data Bank (PDB) code: 4FJO), pseudo-coloured according to amino acid conservation. V, variable residues; C, conserved residues. c, Protein threading model of FAM35A identifies an RPA70-like triple OB-fold architecture. The FAM35A structural model generated using Phyre2 predicts that amino acid residues 429–826 (coloured in green) adopt the same folds as Ustilago maydis RPA7024 (PDB code: 4GOP, chain C; 99.5% confidence in the model23). Blue structural chains from RPA70 are inserted in regions of FAM35A that could not be modelled with high confidence (internal regions and FAM35A residues above 826). In the shieldin complex, RPA34 (opaque red) and RPA14 (opaque yellow) subunits of the heterotrimeric RPA complex might be substituted with SHLD1, SHLD3 and REV7 proteins. d, Pairwise alignment shows 16% sequence identity across human FAM35A isoform 2 (OB-folds 1 to 3) and U. maydis RPA70 (OB-folds 2 to 4; generated with Phyre2). Black and grey shading indicate identical and similar residues, respectively.
Extended Data Fig. 6 SHLD3 (FLJ26957) mediates 53BP1-dependent NHEJ.
a, Immunoblot showing levels of REV7, 53BP1 and RIF1 protein in whole cell lysates prepared from indicated CH12-F3 cell lines. Representative of n = 2 individual experiments. For gel source data, see Supplementary Fig. 1. b, Normal proliferation of stimulated wild type and Shld3−/−, Shld1−/− and Shld2−/− CH12-F3 cells. CFSE dye dilution assay. n = 2 independent experiments. c, Summary of IgM-to-IgA CSR frequencies in indicated control GST or HA–SHLD2(mouse)-complemented Shld2−/− CH12-F3 cells. Data normalized to CSR in wild-type cells. n = 4 independent experiments; mean ± s.d. NC, non-complemented. d, IgM-to-IgA CSR in indicated Shld3−/−CH12-F3 cells ±complementation with wild-type SHLD3 or SHLD3(P53A, P58A) (that is, RBM2 mutated). Data normalized to CSR in wild-type cells. Mean ± s.d. (n = 4). e, Summary of IgM-to-IgA CSR frequencies in Rev7−/− and Rev7−/−Shld3−/− double knockout CH12-F3 clones. Data normalized to CSR in wild-type cells. n = 4 independent experiments; mean ± s.d. f, Histone H3 (control) ChIP efficiencies at indicated Igh and control loci. This panel is related to RPA ChIP data shown in Fig. 4d. Representative data, n = 2 independent experiments, mean ± s.d., 2 or 3 qPCR replicates. g, Indicated CH12-F3 lines stimulated with anti-CD40 antibody, IL-4 and TGFβ1 (30 h) were subjected to ChIP experiments with RPA34 and histone H3 (control) antibodies. Representative data, n = 2 independent experiments, mean ± s.d., 2 or 3 qPCR replicates. h, Cell cycle of indicated CH12-F3 lines after MMC treatment. Proportions (%) of sub-G1, G1, S and G2/M events. n = 3 independent experiments; mean ± s.d. i, Change (%) of Cas9-dependent indels at the indicated sgRNA locus in KB1P-G3 cells after outgrowth in DMSO or olaparib (300 nM) for 7 days. Representative data, 2 independent experiments. j, As in i but with 53bp1−/− KB1P-G3 cells. Representative data, n = 2 independent experiments. k, As in Fig. 4e but with 53bp1−/− KB1P-G3 cells. Representative data, n = 3 independent experiments.
Supplementary information
Supplementary Figures
This file contains the uncropped blots, scans, and FACS gating strategy 1-2.
Rights and permissions
About this article
Cite this article
Ghezraoui, H., Oliveira, C., Becker, J.R. et al. 53BP1 cooperation with the REV7–shieldin complex underpins DNA structure-specific NHEJ. Nature 560, 122–127 (2018). https://doi.org/10.1038/s41586-018-0362-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0362-1
This article is cited by
-
Multifaceted roles of CCAR family proteins in the DNA damage response and cancer
Experimental & Molecular Medicine (2024)
-
The complementarity of DDR, nucleic acids and anti-tumour immunity
Nature (2023)
-
Targeting DNA damage response pathways in cancer
Nature Reviews Cancer (2023)
-
Leveraging the replication stress response to optimize cancer therapy
Nature Reviews Cancer (2023)
-
Shieldin complex assembly kinetics and DNA binding by SHLD3
Communications Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.