Abstract
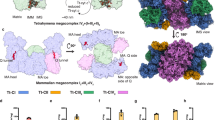
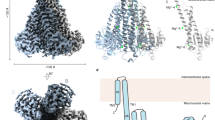
The mitochondrial calcium uniporter (MCU) is a highly selective calcium channel and a major route of calcium entry into mitochondria. How the channel catalyses ion permeation and achieves ion selectivity are not well understood, partly because MCU is thought to have a distinct architecture in comparison to other cellular channels. Here we report cryo-electron microscopy reconstructions of MCU channels from zebrafish and Cyphellophora europaea at 8.5 Å and 3.2 Å resolutions, respectively. In contrast to a previous report of pentameric stoichiometry for MCU, both channels are tetramers. The atomic model of C. europaea MCU shows that a conserved WDXXEP signature sequence forms the selectivity filter, in which calcium ions are arranged in single file. Coiled-coil legs connect the pore to N-terminal domains in the mitochondrial matrix. In C. europaea MCU, the N-terminal domains assemble as a dimer of dimers; in zebrafish MCU, they form an asymmetric crescent. The structures define principles that underlie ion permeation and calcium selectivity in this unusual channel.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Deluca, H. F. & Engstrom, G. W. Calcium uptake by rat kidney mitochondria. Proc. Natl Acad. Sci. USA 47, 1744–1750 (1961).
Montero, M. et al. Chromaffin-cell stimulation triggers fast millimolar mitochondrial Ca2+ transients that modulate secretion. Nat. Cell Biol. 2, 57–61 (2000).
Baughman, J. M. et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 (2011).
De Stefani, D., Raffaello, A., Teardo, E., Szabò, I. & Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011).
Chaudhuri, D., Sancak, Y., Mootha, V. K. & Clapham, D. E. MCU encodes the pore conducting mitochondrial calcium currents. eLife 2, e00704 (2013).
Kamer, K. J. & Mootha, V. K. The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 16, 545–553 (2015).
Gunter, K. K. & Gunter, T. E. Transport of calcium by mitochondria. J. Bioenerg. Biomembr. 26, 471–485 (1994).
Kirichok, Y., Krapivinsky, G. & Clapham, D. E. The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427, 360–364 (2004).
Bick, A. G., Calvo, S. E. & Mootha, V. K. Evolutionary diversity of the mitochondrial calcium uniporter. Science 336, 886 (2012).
Sancak, Y. et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342, 1379–1382 (2013).
Perocchi, F. et al. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature 467, 291–296 (2010).
Plovanich, M. et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8, e55785 (2013).
Mallilankaraman, K. et al. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat. Cell Biol. 14, 1336–1343 (2012).
Vais, H. et al. MCUR1, CCDC90A, is a regulator of the mitochondrial calcium uniporter. Cell Metab. 22, 533–535 (2015).
Chaudhuri, D., Artiga, D. J., Abiria, S. A. & Clapham, D. E. Mitochondrial calcium uniporter regulator 1 (MCUR1) regulates the calcium threshold for the mitochondrial permeability transition. Proc. Natl Acad. Sci. USA 113, E1872–E1880 (2016).
Kovács-Bogdán, E. et al. Reconstitution of the mitochondrial calcium uniporter in yeast. Proc. Natl Acad. Sci. USA 111, 8985–8990 (2014).
Tsai, M.-F. et al. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. eLife 5, e15545 (2016).
Oxenoid, K. et al. Architecture of the mitochondrial calcium uniporter. Nature 533, 269–273 (2016).
Moraes, I., Evans, G., Sanchez-Weatherby, J., Newstead, S. & Stewart, P. D. Membrane protein structure determination — the next generation. Biochim. Biophys. Acta 1838 (1 Pt A), 78–87 (2014).
Long, S. B., Tao, X., Campbell, E. B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007).
Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W. A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011).
Wu, J. et al. Structure of the voltage-gated calcium channel Cav1.1 complex. Science 350, aad2395 (2015).
Shen, H. et al. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 355, eaal4326 (2017).
Yan, Z. et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature 517, 50–55 (2015).
Zalk, R. et al. Structure of a mammalian ryanodine receptor. Nature 517, 44–49 (2015).
Fan, G. et al. Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature 527, 336–341 (2015).
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013).
Hille, B. Ionic Channels of Excitable Membranes 3rd edn (Sinauer Associates, Sunderland, 2001).
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 A resolution. Nature 414, 43–48 (2001).
Lee, Y. et al. Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter. EMBO Rep. 16, 1318–1333 (2015).
Lee, S. K. et al. Structural insights into mitochondrial calcium uniporter regulation by divalent cations. Cell Chem. Biol. 23, 1157–1169 (2016).
Miller, A. N. & Long, S. B. Crystal structure of the human two-pore domain potassium channel K2P1. Science 335, 432–436 (2012).
Brohawn, S. G., Campbell, E. B. & MacKinnon, R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516, 126–130 (2014).
Dong, Y. Y. et al. K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac. Science 347, 1256–1259 (2015).
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Kilmartin, J. V., Wright, B. & Milstein, C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93, 576–582 (1982).
Long, S. B., Campbell, E. B. & Mackinnon, R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 (2005).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protocols 9, 2574–2585 (2014).
Molday, R. S. & MacKenzie, D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22, 653–660 (1983).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 184, 226–236 (2013).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protocols 8, 2281–2308 (2013).
Acknowledgements
We thank N. Grigorieff, members of his laboratory, and the staff at the Howard Hughes Medical Institute Cryo-EM facility for training in cryo-EM; R. K. Hite and members of the Long laboratory for discussions; the staff of the New York Structural Biology Center Simons Electron Microscopy Center, M. Ebrahim, and M. J. de la Cruz of the Memorial Sloan Kettering Cancer Center Cryo-EM facility for help with data collection; and J. Goldberg for spectrophotometer use. This work was supported, in part, by an NIH core facilities grant to Memorial Sloan Kettering Cancer Center (P30 CA008748), by an NIH Medical Scientist Training Program grant (T32GM007739 for A.F.S.), and by an NIH grant (R01GM094273 to S.B.L.).
Reviewer information
Nature thanks P. Yuan and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
R.B. and C.W. performed cryo-EM studies of CeMCU and zebrafish MCU, respectively, and other experiments. A.F.S. developed MCU and EMRE double-knockout cells. S.B.L. directed the research and assisted with cryo-EM. All authors contributed to data analysis and the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Structure-based sequence alignment.
The amino acid sequences of C. europaea, Pyronema omphalodes, D. discoideum, C. elegans, zebrafish (D. rerio) and human MCUs are aligned and coloured according to the ClustalW convention (UniProt accession numbers: W2SDE2, U4LFM6, Q54LT0, Q21121, Q08BI9 and Q8NE86, respectively). The secondary structure is indicated with ribbons representing α-helices, solid lines representing structured loop regions, and dashed lines representing disordered regions. The WDXXEP signature sequence is highlighted with a red line.
Extended Data Fig. 2 Functional analysis of MCU.
a, b, Representative Ca2+ uptake experiments using digitonin-permeabilized MCU/EMRE knockout cells without transfection (red line), or expressing zebrafish MCU and EMRE (black line), or expressing zebrafish MCUΔNTD and EMRE (grey line). Blue arrows indicate additions of 5 μM CaCl2. A decrease in fluorescence following addition of Ca2+ is indicative of Ca2+ uptake (for example, black and grey traces). Ruthenium red (RuRed), a proton ionophore (CCCP), or a Ca2+ ionophore (ETH129) were added as controls towards the end of each experiment as indicated. RuRed prevents Ca2+ uptake, ETH129 demonstrates that the mitochondria are intact, and CCCP demonstrates that Ca2+ uptake is dependent upon the proton gradient. Analogous distinct experiments were repeated a total of six times and yielded similar results. c, Rho-1D4 western blot analysis of the cell lysates from a, demonstrating expression of MCU and EMRE, which were C-terminally tagged with a 1D4 peptide (see Methods). Repeated twice with similar results. d, f, Representative mitochondrial Ca2+ uptake experiments using MCU/EMRE knockout cells for the indicated mutations of human MCU when co-transfected with EMRE (from which the data shown in Fig. 4d are derived, see Methods). Independent experiments were repeated with similar results: n = 8 for wild type, n = 9 for M263A, and n = 3 for the remainder of the mutants. e, g, Full-size western blots of the cell lysates from d and f showing protein expression levels (corresponding to Fig. 4d; detected using a Rho-1D4 antibody). Repeated twice with similar results. h–j, Subunit stoichiometry analysis of MCU proteins using crosslinking. h, Crosslinking of purified CeMCU in the detergent n-dodecyl-β-d-maltoside (DDM). Indicated concentrations of the crosslinker bis(sulfosuccinimidyl) suberate (BS-3) were incubated with purified CeMCU. Analysis is by SDS–PAGE and coomassie stain. Molecular weight standards are located in the first lane and their positions indicated. The calculated molecular weight of CeMCU is 39.6 kDa based upon its amino acid sequence. Repeated three times with similar results. i, Crosslinking of human MCU expressed in HEK293 membranes. Indicated concentrations of the membrane-permeable crosslinker disuccinimidyl glutarate (DSG) were used and human MCU was detected by western blot using a C-terminal Rho-1D4 antibody tag (Methods). Molecular weight standards are located in the first lane and their positions indicated. We note that some oligomerization of human MCU was observed without crosslinker; this phenomenon has been observed previously for human MCU32. Repeated four times with similar results. j, Crosslinking of purified zebrafish MCU in the detergent digitonin. Indicated concentrations of BS-3 crosslinker were used. Samples were analysed by SDS–PAGE using coomassie stain. Molecular weight standards are located in the last lane and their positions are indicated. Asterisks indicate protein impurities. Repeated twice with similar results.
Extended Data Fig. 3 Flowchart for cryo-EM data processing of CeMCU.
Details can be found in the Methods.
Extended Data Fig. 4 Flowchart for cryo-EM data processing of zebrafish MCU.
a, Initial model generation and improvement. b, 3D refinement using the improved initial model from a. Details can be found in the Methods.
Extended Data Fig. 5 Cryo-EM reconstruction of zebrafish MCU.
a, Orthogonal slices (top) and orthogonal 2D projections (bottom) of the final 3D reconstruction (from cisTEM). b, Angular orientation distribution of the particles used in the final reconstruction. The particle distribution is indicated by different colour shades. c, Gold-standard FSC curve of the final 3D reconstruction. The resolution is ~8.5 Å at the FSC cutoff of 0.143. A thin vertical line indicates that only spatial frequencies to 1/(15 Å) were used to determine particle alignment parameters during refinement in cisTEM.
Extended Data Fig. 6 Cryo-EM structure determination and density of CeMCU.
a, Orthogonal slices (top) and orthogonal 2D projections (bottom) of the final 3D reconstruction (from cisTEM). b, Angular orientation distribution of the particles used in final reconstruction. The particle distribution is indicated by different colour shades. c, Gold-standard FSC curve of the final 3D reconstruction. The resolution is 3.2 Å at the FSC cutoff of 0.143. A thin vertical line indicates that only spatial frequencies to 1/(7 Å) were used to determine particle alignment parameters during refinement. d, Local resolution of the map estimated using the blocres program and coloured as indicated. e, Model validation. Comparison of the FSC curves between model and half map 1 (work), model and half map 2 (free), and model and full map are plotted in green, red and blue, respectively. f–i, Densities (8σ contours) of α-helical regions of CeMCU are shown in the context of the atomic model with side chains shown as sticks and the backbone as ribbons. j, Stereo representation of the density (8σ contour) of the NTDs, with the interface between chains A and B in the foreground.
Extended Data Fig. 7 Structure of CeMCU.
a, Ribbon representation of an isolated subunit from Fig. 1c, viewed parallel to the membrane, with the structural features labelled. Disordered regions connecting TM2 and α6 are indicated as dashed lines. b, c, IMS and matrix views of Fig. 1c. d, e, Molecular surface coloured according to electrostatic potential (red, −8 kT e−1; white, neutral; blue, +8 kT e−1). d, Shown in the same orientation as Fig. 1c with approximate membrane boundaries as grey bars. e, IMS view. A green sphere indicates the position of Ca2+ in site 1. f, Overall structure of CeMCU, depicted similarly to Fig. 1. g, Subunit interactions within the TMD. Ribbon representations of TM1 and TM2 from one subunit (green) and TM2 of the neighbouring subunit (red) are shown. Residues participating in van der Waals or hydrogen bonding interactions are shown as sticks; dashed lines indicate hydrogen bonds. Atom colouring: nitrogen, blue; oxygen, red; and sulfur, green. h, Interfaces within the NTDs. The NTDs of subunit A, B and C (green, red and yellow, respectively) are shown in ribbon and surface representations. Each NTD contains six β-strands (β1 through β6) and two α-helices (α1 and α2). There are four interfaces between the NTDs, and these are of two types. Interface 1 (for example, between protomers A and B or C and D in the atomic model) consists of an interaction between β3 of one NTD and the β1–β2 loop of another; this interface buries 2,188 Å2 of molecular surface in the assembled channel. Interface 2 (for example, between protomers A and D or B and C) is less extensive, burying 1,440 Å2 total surface area, and involves contacts located near the N-terminal ends of the α1 helices from two NTDs. i, j, Details of interfaces 1 and 2 between NTDs, respectively.
Extended Data Fig. 8 Comparison with an NMR structure (PDB: 5ID3).
Left, various representations of the cryo-EM structure of CeMCU. The NTD, TM1ext, TM1, TM2 and coiled-coil regions are shown in different colours. Right, structure deduced from NMR studies of C. elegans MCU-ΔNTD (PDB: 5ID3). It is coloured according to the cryo-EM structure using the sequence alignment shown in Extended Data Fig. 1. The boxed regions highlight differences in vicinity of the WDXXEP signature sequence and pore. Pore-lining residues (D225, E228, T231 and Y232; C. europaea numbering) are shown as sticks.
Extended Data Fig. 9 Structural comparison of the NTDs of CeMCU, zebrafish MCU and human MCU.
a, An NTD from the cryo-EM structure of CeMCU (magenta) is superimposed with the crystal structure of an isolated NTD from human MCU (grey, PDB: 5KUG, r.m.s.d. = 2.0 Å). The secondary structure features are indicated; two views are shown. b, Crystal lattice from an X-ray structure of an isolated NTD of human MCU (PDB: 4XTB). Four neighbouring NTDs are coloured and other NTDs are grey. A similar arrangement is present in PDB 5KUG. c, Matrix view of the cryo-EM reconstruction of zebrafish MCU (from Fig. 2d).
Supplementary information
Rights and permissions
About this article
Cite this article
Baradaran, R., Wang, C., Siliciano, A.F. et al. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature 559, 580–584 (2018). https://doi.org/10.1038/s41586-018-0331-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0331-8
This article is cited by
-
Pulmonary hypertension
Nature Reviews Disease Primers (2024)
-
Structural and functional analysis of human pannexin 2 channel
Nature Communications (2023)
-
The Role of PGC-1α-Mediated Mitochondrial Biogenesis in Neurons
Neurochemical Research (2023)
-
MiR-25 overexpression inhibits titanium particle-induced osteoclast differentiation via down-regulation of mitochondrial calcium uniporter in vitro
Journal of Orthopaedic Surgery and Research (2022)
-
Mitochondrial calcium uniporter stabilization preserves energetic homeostasis during Complex I impairment
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.