Abstract
Mammalian cells integrate mitogen and stress signalling before the end of G1 phase to determine whether or not they enter the cell cycle1,2,3,4. Before cells can replicate their DNA in S phase, they have to activate cyclin-dependent kinases (CDKs), induce an E2F transcription program and inactivate the anaphase-promoting complex (APC/CCDH1, also known as the cyclosome), which is an E3 ubiquitin ligase that contains the co-activator CDH1 (also known as FZR, encoded by FZR1). It was recently shown that stress can return cells to quiescence after CDK2 activation and E2F induction but not after inactivation of APC/CCDH1, which suggests that APC/CCDH1 inactivation is the point of no return for cell-cycle entry3. Rapid inactivation of APC/CCDH1 requires early mitotic inhibitor 1 (EMI1)3,5, but the molecular mechanism that controls this cell-cycle commitment step is unknown. Here we show using human cell models that cell-cycle commitment is mediated by an EMI1–APC/CCDH1 dual-negative feedback switch, in which EMI1 is both a substrate and an inhibitor of APC/CCDH1. The inactivation switch triggers a transition between a state with low EMI1 levels and high APC/CCDH1 activity during G1 and a state with high EMI1 levels and low APC/CCDH1 activity during S and G2. Cell-based analysis, in vitro reconstitution and modelling data show that the underlying dual-negative feedback is bistable and represents a robust irreversible switch. Our study suggests that mammalian cells commit to the cell cycle by increasing CDK2 activity and EMI1 mRNA expression to trigger a one-way APC/CCDH1 inactivation switch that is mediated by EMI1 transitioning from acting as a substrate of APC/CCDH1 to being an inhibitor of APC/CCDH1.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Arora, M., Moser, J., Phadke, H., Basha, A. A. & Spencer, S. L. Endogenous replication stress in mother cells leads to quiescence of daughter cells. Cell Reports 19, 1351–1364 (2017).
Barr, A. R. et al. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 8, 14728 (2017).
Cappell, S. D., Chung, M., Jaimovich, A., Spencer, S. L. & Meyer, T. Irreversible APCCdh1 inactivation underlies the point of no return for cell-cycle entry. Cell 166, 167–180 (2016).
Yang, H. W., Chung, M., Kudo, T. & Meyer, T. Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature 549, 404–408 (2017).
Hsu, J. Y., Reimann, J. D., Sørensen, C. S., Lukas, J. & Jackson, P. K. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APCCdh1. Nat. Cell Biol. 4, 358–366 (2002).
Ferrell, J. E. Jr. Bistability, bifurcations, and Waddington’s epigenetic landscape. Curr. Biol. 22, R458–R466 (2012).
Choudhury, R. et al. APC/C and SCFcyclin F constitute a reciprocal feedback circuit controlling S-phase entry. Cell Reports 16, 3359–3372 (2016).
Fukushima, H. et al. SCF-mediated Cdh1 degradation defines a negative feedback system that coordinates cell-cycle progression. Cell Reports 4, 803–816 (2013).
Lukas, C. et al. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature 401, 815–818 (1999).
Huang, X. et al. Deubiquitinase USP37 is activated by CDK2 to antagonize APCCDH1 and promote S phase entry. Mol. Cell 42, 511–523 (2011).
Miller, J. J. et al. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 20, 2410–2420 (2006).
Margottin-Goguet, F. et al. Prophase destruction of Emi1 by the SCFβTrCP/Slimb ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell 4, 813–826 (2003).
Lindon, C. & Pines, J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell Biol. 164, 233–241 (2004).
Zeng, X. et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18, 382–395 (2010).
Rape, M., Reddy, S. K. & Kirschner, M. W. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell 124, 89–103 (2006).
Meyer, H. J. & Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 (2014).
Moshe, Y., Bar-On, O., Ganoth, D. & Hershko, A. Regulation of the action of early mitotic inhibitor 1 on the anaphase-promoting complex/cyclosome by cyclin-dependent kinases. J. Biol. Chem. 286, 16647–16657 (2011).
Frye, J. J. et al. Electron microscopy structure of human APC/CCDH1–EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat. Struct. Mol. Biol. 20, 827–835 (2013).
Chang, L. F., Zhang, Z., Yang, J., McLaughlin, S. H. & Barford, D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature 513, 388–393 (2014).
Wang, W. & Kirschner, M. W. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat. Cell Biol. 15, 797–806 (2013).
Dimova, N. V. et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat. Cell Biol. 14, 168–176 (2012).
Thrower, J. S., Hoffman, L., Rechsteiner, M. & Pickart, C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 (2000).
Spencer, S. L. et al. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013).
Sherr, C. J. Cancer cell cycles. Science 274, 1672–1677 (1996).
Song, L. & Rape, M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol. Cell 38, 369–382 (2010).
Pierce, N. W., Kleiger, G., Shan, S. O. & Deshaies, R. J. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 (2009).
Acknowledgements
We thank M. Chung, C. Liu, A. Hayer, H. W. Yang, L. Daigh, Y. Fan, A. Jaimovich, M. Köberlin, S. Spencer, S. Collins, N. Ratnayeke, J. Kamenz, and J. Ferrell for helpful discussions, and the Stanford Shared FACS Facility for cell sorting. S.D.C. was supported by the Damon Runyon Cancer Research Foundation (DRG-2141) and by the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute. K.G.M., D.G. and L.R.P. were supported by NIH F32 Ruth L. Kirschstein fellowships (GM120956, GM116328 and GM125246, respectively). M.R. is supported by NIGMS R01 grants (GM083064) and is an Investigator of the Howard Hughes Medical Institute. T.M. is supported by NIGMS R01 grants (R35GM127026, GM118377 and GM063702).
Reviewer information
Nature thanks C. Tang and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Competing interests:
M.R. is a co-founder and consultant to Nurix, a biotech company in the ubiquitin space.
Author information
Authors and Affiliations
Contributions
S.D.C. designed and carried out most experiments. K.G.M. performed in vitro ubiquitination assays. D.G. performed synchronization and western blot experiments. L.R.P. made lentivirus and stable cell lines. M.R. supervised the in vitro ubiquitination studies. S.D.C. and T.M. conceived the project, analysed the data and wrote the manuscript. All authors discussed the results and the manuscript. T.M. supervised the study.
Corresponding authors
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
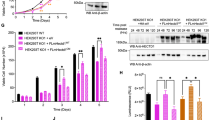
Extended Data Fig. 1 Single-cell APC/CCDH1 data used to determine hysteresis curves.
a, Experimental setup for Fig. 1b, d. and Extended Data Fig. 1b. Asynchronous MCF10A cells were tracked and imaged for approximately 18 h and then treated with either DMSO or the CDK1/2 inhibitor (at concentrations indicated in b–d). Two subpopulations were analysed: cells in G1 phase (drug added while APC/CCDH1 on) or cells in S/G2 phase (drug added while APC/CCDH1 off). b, Single-cell traces of APC/C activity in MCF10A cells treated with DMSO or 30 μM CDK1/2 either in G1 phase (top and middle) or S phase (bottom). Traces were coloured red if APC/CCDH1 ended up inactive and traces were coloured black if APC/CCDH1 ended up active, at 5 h after the addition of the drug. (n = 200, 135 and 197 cells in top, middle and bottom panel, respectively). Histograms to the right of each panel give APC/C activity at the last frame of the movie for each cell. Pie chart insets give the percentage of cells with APC/C still active at the end of the movie. c, Scatter plot of CDK2 activity at the time of drug spike, plotted against the CDK2 activity from the same cell 1 h later. Cells at all phases of the cell cycle were included. d, Top, representative APC/C activity traces for MCF10A cells in G1 phase at time of the indicated treatment. Bottom, representative APC/C activity traces for MCF10A cells in S/G2 phase at time of the indicated treatment. Traces were coloured red if APC/CCDH1 was inactive and traces were coloured black if APC/CCDH1 was still active, at 5 h after addition of the drug. Pie charts show the percentage of cells with APC/CCDH1 still active, used to make the dose–response curve in Fig. 1b. e, Table of the EC50 for the dose–response curves in Fig. 1b, d ± s.e.m. n = 3 independent experiments. Doses above 30 μM CDK1/2 inhibitor killed the cells, making it impossible to calculate an EC50 above 30 μM. f, MCF10A cells were treated with 10 μM CDK1/2 inhibitor and binned by their APC/CCDH1 activity at the time of drug addition. The percentage of cells in each bin with APC/CCDH1 activity on at 2.5 h after drug addition was calculated. Data were analysed by nonlinear regression (sigmoidal dose–response, variable slope). Error bars are s.d. from two independent experiments. g, Scatter plot of APC/C activity at the time of drug spike, plotted against the APC/C activity from the same cell 2.5 h later. Vertical dashed line represents the 70% inactive threshold. Horizontal line indicates the point at which APC/C activity is below 0.1, and is considered to be off.
Extended Data Fig. 2 siRNA validation experiments.
a–e, HeLa (a, b, e) or U2OS (c, d) cells were treated with the indicated siRNA for 24 h before fixation and immunostaining. Scale bars, 20 μm. Single-cell quantification of immunofluorescence data was used to generate the histograms. In a, n = 7,152 cells for siControl and n = 4,051 cells for siCCNA2. In b, n = 7,772 cells for siControl and n = 3,916 cells for siSKP2. In c, n = 7,003 cells for siControl and n = 8,589 cells for siCCNF. In d, n = 9,393 cells for siControl and n = 7,871 cells for siEMI1. e, n = 64,574 cells for siControl and n = 53,338 cells for siCCNE1 and siCCNE2.
Extended Data Fig. 3 EMI1 is responsible for rapid APC/CCDH1 inactivation in multiple cell lines.
a, b, Single-cell traces of APC/C activity computationally aligned to the point at which APC/CCDH1 first starts to inactivate, in cells treated with the indicated siRNA 6 h before the start of imaging. Black line, median APC/C activity trace. Panels on the far right show median APC/C activity trace ± s.d. HeLa cells, n = 129 (representative traces plotted)/602 (all traces used to calculate median), 126/384, 124/399, 120/228, 138/400 cells with siControl, siCCNA2, siSKP2, siCCNF and siEMI1, respectively (a); MCF10A cells, n = 139/400, 130/400, 134/400 cells with siControl, siCCNF and siEMI1, respectively (b). c, Single-cell traces of APC/C activity computationally aligned to mitosis in U2OS cells treated with the indicated siRNA 6 h before the start of imaging. Black line, median APC/C activity trace. Traces in U2OS cells had to be aligned by mitosis because APC/CCDH1 did not inactivate when cells were treated with siRNA targeting EMI1. n = 200 representative cells per condition out of a total of 2,068, 1,531 or 875 cells for siControl, siCCNF and siEMI1, respectively. d, Single-cell traces of APC/C activity in MCF10A cells treated EMI1 siRNA. At the indicated time in S phase, 1 μM CDK1/2 inhibitor was spiked in. Traces were coloured red if APC/CCDH1 stayed inactive and traces were coloured black if APC/CCDH1 turned back on, 5 h after addition of the drug. n = 80. Inset: pie chart of the percentage of cells that turned APC/C back on at the end of the movie. e, Schematic of proposed bistable switch involving cyclin E1– and cyclin E2–CDK2, APC/C and EMI1.
Extended Data Fig. 4 Measuring protein and mRNA dynamics from fixed cells.
a, HeLa cells were synchronized with thymidine followed by nocodazole and mitotic shake-off (see Fig. 2a). After shake-off, cells were immediately plated in a 96-well plate for live-cell imaging. Top, single-cell traces of APC/C-degron levels. Black line, median APC/C-degron level. Bottom, single-cell APC/C activity traces. Dashed line indicates the time at which APC/CCDH1 first starts to inactivate. n = 200 representative cells out of a total of 3,190 cells used to calculate the median. b, Asynchronous HeLa cells were exposed to 10 μM EdU for 15 min, fixed and then stained for both EMI1 and EdU incorporation. Top, density scatter plot of DNA content versus EdU levels, used to gate cells into specific cell-cycle phases. (for example, G1 cells have 2n DNA and low EdU, S phase cells have greater than 2n DNA and high EdU and so on). Cells were designated as early S phase if they had 2n DNA and sub-maximal EdU, which indicates they were in S phase for less than 15 min at the time of fixation. Bottom, density scatter plot of DNA content versus EMI1 levels. Because we measured EdU and EMI1 in the exact same cells, cell-cycle phase gates were applied to the EMI1 scatter plot. Each cell was coloured based on the cell-cycle phase that cell was in. Large dots with a black border mark the median values for each gated population. The single-component histograms for EMI1 levels in each cell-cycle phase are shown on the right. c, HeLa Cells were treated as in Fig. 2a, but 2 h after release, cells were treated with either DMSO or 10 μM MG132. Lysates were collected 2 h later (a total of 4 h after release). Image is representative of n = 3 independent experiments. d, Schematic of the regulation of three key G1 and S phase genes, GMNN, EMI1 and CCNE1, by both transcription and ubiquitination-mediated degradation. e, HeLa cells were imaged and tracked for 20 h, and the time each cell went through anaphase relative to the end of the movie was recorded. Cells were then fixed and stained with single molecule mRNA FISH probes for the indicated genes (top), or with antibodies against the indicated proteins (bottom). Data are a scatter plot of the single-cell mRNA or protein levels as a function as the time relative to mitosis. Solid line represents the median levels from 1-h bins. n = 2 independent experiments. f, Cells treated as in e, but before fixation cells were incubated with 100 μM EdU for 5 min. Dashed line indicates the first bin in which EdU incorporation goes above background, which indicates that cells have entered the S phase. Data are from one representative experiment. For the panel on the right, single-cell data were binned in 1-h intervals and the median ± s.e.m. was plotted. n = 6 replicates.
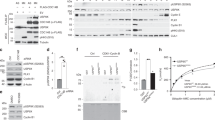
Extended Data Fig. 5 EMI1 accumulates coincident with APC/CCDH1 inactivation.
a–c, Normalized mRNA and protein levels for the indicated genes. Data from mRNA and protein are plotted on the same x axis to enable comparison of the kinetics. The start of S phase was determined by EdU incorporation (see Extended Data Fig. 4e). Data are median value for each bin. n = 2 independent experiments. d, Density scatter plot of EMI1 protein levels and geminin protein levels measured in the same single cells. Single-component histograms are shown as insets. e, Density scatter plot of cyclin E1 protein levels and geminin protein levels measured in the same single cells. Single-component histograms are shown as insets. f, HeLa cells were treated with either control siRNA (black line) or CDH1 siRNA (coloured line) and then imaged and tracked for 20 h. The time at which each cell went through anaphase relative to the end of the movie was recorded. Cells were then fixed and immunofluorescence was performed with antibodies to the indicated proteins. The time required to reach half-maximal protein abundance is indicated with a dashed line (labelled t1/2). Data are median values ± s.e.m. and the line of best fit was calculated using nonlinear regression. Data were normalized to maximum value. n = 8 independent experiments. g, Bar graph of the difference in t1/2 times in cells treated with control siRNA and CDH1 siRNA from j. Individual data points are shown as dots. Error bars are s.e.m. P values from one-way ANOVA with Tukey multiple comparisons test. n = 8 independent experiments. h, i, In vitro ubiquitination time course using immuno-purified APC/C and either recombinant EMI1 (h) or geminin (i). Bottom, lighter exposure showing the un-modified substrate. n = 1 experiment. j, In vitro ubiquitination assay using immuno-purified APC/CCDH1, recombinant EMI1 and the indicated ubiquitin mutant. E2C/S, E2 enzymes UBE2C and UBE2S. n = 1 experiment.
Extended Data Fig. 6 EMI1 and geminin protein levels are tightly correlated during the cell cycle.
a, Model of APC/CCDH1 architecture. For simplicity, the substrate-binding region comprising primarily CDH1 and APC10 is designated as site 1, and the E2 ubiquitin-conjugating enzymes UBCH10 and UBE2S are designated as site 2. Sub, substrate; Ub, ubiquitin. b, Regulation of APC/C by wild-type EMI1. EMI1 is thought to bind both to site 1 and to site 2 to inhibit APC/C. c, d, Regulation of APC/C by two EMI1 mutants. Substrates are recruited to APC/C through a substrate-binding region (site 1). Ubiquitin is transferred to the substrate by UBCH10 and UBE2S (site 2). e, Signalling diagram of the G1 cell-cycle program, showing how inhibiting CDK4/6 activity with a small molecule inhibitor will prevent APC/CCDH1 inactivation. f, MCF10A cells expressing either empty vector or various EMI1 variants were treated with DMSO. Single-cell APC/C activity traces were computationally aligned to the time of mitosis. DMSO treatment was administered before mitosis, as indicated by the arrow. n = 120 cells for each condition. g, Median levels of APC/C degron (red) or wild-type EMI1 (EMI1WT; green) stably expressed in the same MCF10A cells. n = 330 cells. mit, mitosis. h, Median expression levels of ectopically expressed EMI1 mutants tagged with mCitrine. Measurments for each single-cell were the maximum fluorescence detected during each cell cycle. Single-cell data were then averaged for each replicate. Individual data points are shown as dots. Data are median values ± s.d. n = 3 independent experiments. i, Left, median APC/C-degron levels measured over 24 h in MCF10A cells. Arrows indicate mitosis. n = 88,125 cells. Right, median levels of APC/C degron (red) or EMI1(C401S) (green) stably expressed in the same cells. n = 3,618 cells. Inset diagram shows that EMI1(C401S) is a substrate of APC/CCDH1 and not an inhibitor. j, Single-cell traces of APC/C degron (red) or EMI1(C401S) (green) levels in MCF10A cells. Six representative cells from i are shown. k, Density scatter plot of APC/C-degron levels versus EMI1(C401S) levels measured in the same cells. l, Percentage of cells from Fig. 3b–e that inactivated their APC/CCDH1 at least 5 h after mitosis. n = 3 biological repeats. Individual data points are shown as dots. Error bars are s.d.
Extended Data Fig. 7 Mathematical model for an EMI1–APC/C dual-negative feedback loop.
a, Differential equation for the regulation of EMI1 protein concentration. Constant synthesis (S) is assumed for the steady-state model. The model includes reversible low-affinity binding of EMI1 to the substrate-binding site (γ+ or γ−) and inhibitory binding that blocks ubiquitin chain elongation, but not mono-ubiquitination. EMI1 ubiquitination is assumed to be distributive, requiring five ubiquitin elongations to become fully ubiquitinated for effective degradation. APC/CCDH1 activity is inhibited by CDK2 activity or by EMI1 binding to the inhibitory site, allowing EMI1 to be subsequently de-ubiquitinated and stabilized. b, A single-cell APC/C activity and CDK2 activity trace. Each steady state from Fig. 3g is marked. At the indicated time, 3 μM of CDK1/2 inhibitor was added but APC/C activity remains off, as predicted in Fig. 3g.
Extended Data Fig. 8 EMI1 is an APC/C substrate at low concentrations but an inhibitor of ubiquitin chain elongation at high concentrations.
a, In vitro ubiquitination reaction of recombinant geminin using immuno-purified APC/C and a range of concentrations of recombinant EMI1. Non-specific bands are marked with *. n = 1 experiment. b, In vitro ubiquitination time course using immuno-purified APC/C and 2,944 nM recombinant EMI1. Only mono-ubiquitinated EMI1 was observed after 30 min. n = 1 experiment. c, In vitro ubiquitination assay using immuno-purified APC/CCDC20 from cells arrested in mitosis and either 23 nM or 2,944 nM recombinant EMI1. After the in vitro reaction, equal amounts of EMI1 were loaded onto the gel for western blotting. n = 1 experiment. d, Quantification of fully ubiquitinated EMI1 from Fig. 4b using densitometry. Data are plotted as the percentage of total EMI1. The line of best fit was determined using the Hill equation. Dashed lines indicate an EC50 of about 175 nM. e, Steady-state analysis of EMI1 levels as a function of the degradation and synthesis rate. Assuming constant EMI1 synthesis (dashed lines), there are two stable steady states (red and blue dots). The system can transition to a mono-stable state with APC/C off and high levels of EMI1 by increasing the EMI1 synthesis rate above a threshold level (red arrow). Conversely, the system can transition to a different mono-stable state with APC/C on and low levels of EMI1 by decreasing the EMI1 synthesis rate (blue arrow). This system also exhibits hysteresis, as the occupied steady state depends on the previously occupied state (red dots versus blue dots).
Extended Data Fig. 9 Cyclin E is dispensable for APC/CCDH1 inactivation in cells with constitutively active E2F.
a, b, Single-cell traces of APC/C activity computationally aligned to mitosis in either HeLa (n = 200 cells per condition out of a total of 1,284, 434 and 462 cells for siControl, siCCNE1 and siCCNE2, and siEMI1, respectively, used to calculate the median and s.d.) (a) or U2OS (n = 200 cells per condition out of a total of 551, 300 and 271 cells for siControl, siCCNE1 and siCCNE2, and siEMI1, respectively, used to calculate the median and s.d.) (b) cells treated with the indicated siRNA 6 h before the start of imaging. Black line, median APC/C activity trace. Bottom graph in each panel shows median APC/C activity trace ± s.d. c, U2OS cells were treated with either control, CCNE1 and CCNE2, or EMI1 siRNA for 4 h before the start of time-lapse imaging. APC/C and CDK2 activity were measured in the same cells, and traces were aligned to mitosis. Data are median activity trace ± s.e.m. n = 200 cells. d, The time between anaphase and at which relative APC/C activity reaches below 0.2 was calculated for each cell from Fig. 4f. Times from all cells were averaged for each replicate. Individual data points are shown as dots. Data are median times ± s.e.m. P values from one-way ANOVA with Tukey multiple comparisons test. n = 3 independent experiments. e, Single-cell APC/CCDH1 inactivation slope at the G1–S transition for each cell from Fig. 4f. Slopes from all cells were averaged for each replicate. Data are median APC/CCDH1 inactivation slopes ± s.e.m. P values from one-way ANOVA with Tukey multiple comparisons test. Individual data points are shown as dots. n = 3 independent experiments. f, Histogram of the time between CDK2 activation and APC/CCDH1 inactivation in HeLa cells treated with the indicated siRNA. Values above zero indicate CDK2 turns on before APC/CCDH1 turns off, and values below zero indicate APC/CCDH1 turns off before CDK2 turns on.
Extended Data Fig. 10 APC/CCDH1 inactivates before CDK2 activation in cells with cyclin E1 and cyclin E2 knocked down.
a, b, Single-cell traces of APC/C (red) and CDK2 (green) activity in HeLa cells treated with either control siRNA (a) or CCNE1 and CCNE2 siRNA (b). Ten representative cells are shown for each condition. Black arrow, time of mitosis; green dashed line, time at which CDK2 activity first starts to rise; red dashed line, time at which active APC/C first starts to inactivate. c, Phase-plane diagram of median CDK2 and APC/C activity measured simultaneously in the same U2OS cells over time, starting in mitosis or G0 phase and progressing to S phase. n = 270 cells per condition. d, Scheme of the regulation of APC/CCDH1 inactivation in the presence (wild type) or absence of cyclin E. Cells can bypass the requirement for cyclin E1– and cyclin E2–CDK2 in G1 phase by upregulating EMI1 mRNA. HeLa and USO2 cells bypass cyclin E1– and cyclin E2–CDK2 by having constitutively active E2F1–3.
Supplementary information
Supplementary Figure 1
This file contains the uncropped gel data for Figures 2 and 4, and Extended Data Figures 4, 5 and 8
Rights and permissions
About this article
Cite this article
Cappell, S.D., Mark, K.G., Garbett, D. et al. EMI1 switches from being a substrate to an inhibitor of APC/CCDH1 to start the cell cycle. Nature 558, 313–317 (2018). https://doi.org/10.1038/s41586-018-0199-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0199-7
This article is cited by
-
FBXO5-mediated RNF183 degradation prevents endoplasmic reticulum stress-induced apoptosis and promotes colon cancer progression
Cell Death & Disease (2024)
-
Effect of Emi1 gene silencing on the proliferation and invasion of human breast cancer cells
BMC Molecular and Cell Biology (2023)
-
BCAS2 regulates granulosa cell survival by participating in mRNA alternative splicing
Journal of Ovarian Research (2023)
-
Commentary: locating the restriction point
Cell Division (2023)
-
Research Progress in Function and Regulation of E3 Ubiquitin Ligase SMURF1
Current Medical Science (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.