Abstract
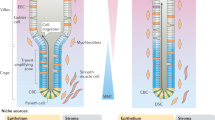
Wnt–β-catenin signalling plays a pivotal role in the homeostasis of the intestinal epithelium by promoting stem cell renewal1,2. In the small intestine, epithelial Paneth cells secrete Wnt ligands and thus adopt the function of the stem cell niche to maintain epithelial homeostasis3,4. It is unclear which cells comprise the stem cell niche in the colon. Here we show that subepithelial mesenchymal GLI1-expressing cells form this essential niche. Blocking Wnt secretion from GLI1-expressing cells prevents colonic stem cell renewal in mice: the stem cells are lost and, as a consequence, the integrity of the colonic epithelium is corrupted, leading to death. GLI1-expressing cells also play an important role in the maintenance of the small intestine, where they serve as a reserve Wnt source that becomes critical when Wnt secretion from epithelial cells is prevented. Our data suggest a mechanism by which the stem cell niche is adjusted to meet the needs of the intestine via adaptive changes in the number of mesenchymal GLI1-expressing cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Valenta, T. et al. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 15, 911–918 (2016).
Fevr, T., Robine, S., Louvard, D. & Huelsken, J. Wnt/β-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559 (2007).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Scadden, D. T. The stem-cell niche as an entity of action. Nature 441, 1075–1079 (2006).
Barker, N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014).
Farin, H. F., Van Es, J. H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529.e7 (2012).
Durand, A. et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl Acad. Sci. USA 109, 8965–8970 (2012).
Kim, T. H., Escudero, S. & Shivdasani, R. A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl Acad. Sci. USA 109, 3932–3937 (2012).
Kabiri, Z. et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 141, 2206–2215 (2014).
Sasaki, N. et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl Acad. Sci. USA 113, E5399–E5407 (2016).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Morikawa, S. et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 206, 2483–2496 (2009).
Kramann, R. et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66 (2015).
Kramann, R. et al. Adventitial MSC-like cells are progenitors of vascular smooth muscle cells and drive vascular calcification in chronic kidney disease. Cell Stem Cell 19, 628–642 (2016).
Bänziger, C. et al. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125, 509–522 (2006).
Degirmenci, B., Hausmann, G., Valenta, T. & Basler, K. Wnt ligands as a part of the stem cell niche in the intestine and the liver. Prog. Mol. Biol. Transl. Sci. 153, 1–19 (2018).
Fafilek, B. et al. Troy, a tumor necrosis factor receptor family member, interacts with Lgr5 to inhibit Wnt signaling in intestinal stem cells. Gastroenterology 144, 381–391 (2013).
van der Flier, L. G., Haegebarth, A., Stange, D. E., van de Wetering, M. & Clevers, H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17 (2009).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Stzepourginski, I. et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl Acad. Sci. USA 114, E506–E513 (2017).
Aoki, R. et al. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell. Mol. Gastroenterol. Hepatol. 2, 175–188 (2016).
Ahn, S. & Joyner, A. L. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505–516 (2004).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Hamilton, T. G., Klinghoffer, R. A., Corrin, P. D. & Soriano, P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol. 23, 4013–4025 (2003).
Sato, T. & Clevers, H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–328 (2013).
Koliaraki, V. & Kollias, G. Isolation of intestinal mesenchymal cells from adult mice. Bio Protoc. 6, e1940 (2016).
Acknowledgements
We thank O. Sansom for providing us with the Gli1-CreERT2 and PdgfraEGFP strains and for suggestions; S. Robine for the Villin-CreERT2 strain; F. Greten, L. Sommer, M. Aguet and G. Christofori for comments; members of the Basler laboratory, in particular C. Cantù, V. S. Salazar and D. Zimmerli, for discussions; E. Escher, E. Tuncer, L. Zurkirchen and V. Parfejevs for technical help; J. Duarte, C. Ewald and A. Henning for help with cell sorting; and C. Aquino and the Functional Genomics Center Zurich for performing scRNA-seq. This work was supported by the Swiss National Science Foundation, the Swiss Cancer League, the University of Zurich Research Priority Program (URPP) ‘Translational Cancer Research’ and the Kanton of Zürich. T.V. is supported by Czech Science Foundation grant 18-21466S and is a fellow of the URPP Translational Cancer Research.
Reviewer information
Nature thanks C. Kuo, L. Samuelson and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
B.D. and T.V. designed and performed experiments and collected and analysed data. T.V. initiated and conceived the research. B.D. and T.V. wrote the manuscript and performed all experiments together with data analysis. S.D. analysed scRNA-seq data. G.H. discussed the data and assisted with manuscript preparation. K.B. initiated and supported the research, discussed the data and assisted with manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
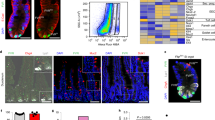
Extended Data Fig. 1 GLI1+ cells are localized adjacent to the base of crypts and are positive for the mesenchymal marker PDGFRA.
a, Sections of duodenal or colonic crypts. Immunohistochemistry for GLI1 (red), E-cadherin (green, top) or ACTA2 (green, bottom) and DAPI (blue) in the duodenum and colon. E-cadherin marks epithelial cells, ACTA2 marks myofibroblasts (mesenchymal cells). b, Cross-sections of duodenal or colonic crypts stained as in a. c, Expression of GLI1 overlaps with that of PDGFRA in mesenchymal cells of Gli1-CreERT2;lox-STOP-lox-tdTomato+;PdgfraEGFP+ mice 24 h after tamoxifen injection. Immunohistochemistry: direct detection of fluorescent proteins in frozen sections. tdTomato (red) marks GLI1+ cells, nuclear EGFP (green) is expressed in PDGFRA+ cells, counterstained with DAPI (blue). d, Intestinal mesenchymal cells isolated from Gli1-CreERT2;lox-STOP-lox-tdTomato+;PdgfraEGFP+ mice 24 h after tamoxifen injection were cultured in Mesencult medium for 5 days. Immunocytochemistry for EGFP (green) marking PDGFRA, tdTomato (red) for GLI1+, DAPI (blue). Full genotype: Gli1-CreERT2;lox-STOP-lox-tdTomatoTg/wt;Pdgfra-EGFPTg/wt; Tg indicates transgenic allele. Scale bar, 50 μm; IMCs, intestinal mesenchymal cells.
Extended Data Fig. 2 GLI1+ cells are persistent in vivo and can act as progenitor cells with potential for tri-lineage differentiation in vitro.
a, tdTomato+ cells are retained in the colon mesenchyme of Gli1-CreERT2;lox-STOP-lox-tdTomato mice 35 days after a single tamoxifen injection. Immunohistochemistry: tdTomato (red) marks GLI1+ cells or their descendants, E-cadherin (green) denotes the shape of crypts, DAPI (blue) stains nuclei. Full genotype: Gli1-CreERT2;lox-STOP-lox-tdTomatoTg/w (Tg indicates transgenic allele). b, Sorted GLI1+(td Tomato+) cells have the capacity to differentiate towards adipocytes (Oil Red O staining), osteoblasts (Alizarin Red S staining) and smooth muscle (ACTA2). c, Undifferentiated GLI1+ (tdTomato+) cells express lower levels of ACTA2 than cells differentiated towards smooth muscle cells. Immunocytochemistry for ACTA2 (green) showing the control (undifferentiated) parallel to b. Scale bar, 50 μm.
Extended Data Fig. 3 GLI1+ cells are essential for the maintenance of the colonic epithelium but dispensable for the normal renewal of the small intestinal (duodenal) epithelium.
a, In the colon, blocking Wnt secretion from GLI1+ cells results in corrupted epithelial morphology in conjunction with loss of proliferation. As no Wnts are secreted from the colonic epithelium, blocking Wnt secretion from both epithelial (Villin-CreERT2 is active in all epithelial cells) and GLI1+ cells is similar to the situation in which Wnt secretion is ablated only in GLI1+ cells. Immunohistochemistry for Ki67 (red), E-cadherin (green) and DAPI (blue) in colon. Days after Cre induction are indicated. b, Normal epithelial morphology of the duodenum in Villin+Gli1-WlscKO animals 12 days after Cre induction. Full genotypes: Gli1-WlscKO (Gli1-CreERT2;Wlsflox/flox), Villin+Gli1-WlscKO (Villin-CreERT2;Gli1-CreERT2;Wlsflox/flox). Scale bar, 50 μm.
Extended Data Fig. 4 Role of Wnt secretion by GLI1+ cells for the maintenance of intestinal epithelial stem cells differs between duodenum and colon.
a, In the duodenum, only simultaneous blocking of Wnt secretion from both the epithelium and GLI1+ cells abrogates the renewal of IES cells marked by LGR5. Immunohistochemistry: LGR5EGFP (green), DAPI (blue), direct detection of fluorescent protein in frozen sections. b, c, In the colon, blocking Wnt secretion from GLI1+ cells results in reduced expression of Wnt target genes and disappearance of IES cell markers (b). Abrogating GLI1-mediated Wnt secretion alters expression of Rspo1 and Rspo3 (c); (real-time RT–qPCR, 12 and 14 days after tamoxifen administration, n = 3 biologically independent mice, mean ± s.d.). d, e, Delivery of external Wnts (WNT3A and WNT2B) delays the loss of expression of the stem cell marker OLFM4 in the duodenal crypts of Villin+Gli1-WlscKO mice. Immunohistochemistry: OLFM4 (red), DAPI (blue). e, External Wnts (WNT3A and WNT2B) prolong the presence of actively proliferating cells and intact crypts within the colonic epithelium of Villin+Gli1-WlscKO mice. Immunohistochemistry: Ki67 (red), E-cadherin (green, marks epithelial cells), DAPI (blue). Scale bars, 50 μm. Full genotypes: Gli1-WlscKO (Gli1-CreERT2;Wlsflox/flox), Villin+Gli1-WlscKO (Villin-CreERT2;Gli1-CreERT2;Wlsflox/flox). Days after Cre induction are indicated.
Extended Data Fig. 5 GLI1+ cells restore the growth of intestinal organoids and maintain the intestinal epithelial stem cells in vitro.
a, GLI1+(tdTomato+) sorted cells change the morphology of duodenal organoids. Small intestinal (duodenal) organoids were cocultured with sorted GLI1+ IMCs. b, c, GLI1+(tdTomato+) sorted cells drive the growth of colonic organoids by providing Wnt ligands. Colonic organoids were cocultured with or without sorted GLI1+ IMCs and grown in medium as indicated. c, GLI1+ (tdTomato+) cells (red) sustain the growth of colonic organoids and small intestinal (duodenal) organoids with ablated Wnt secretion (WlscKO). d, e, Coculture with GLI1+ cells restored the growth of colonic and duodenal WlscKO organoids. The fraction of living organoids after 7 days in culture is shown in the graph. Crypts were seeded at the same initial density. Summarizes data from two independent experiments (n = 2 biologically independent experiments, 3 replicates each); mean ± s.d. Organoids were cultivated in medium containing: ENR = EGF, noggin, R-spondin1; ENRW = EGF, noggin, R-spondin1, WNT3A. IMCsGLI1 are sorted GLI1+ IMCs. Scale bar, 50 μm. f, Scheme depicting how GLI1+ (tdTomato+) cells were sorted. g, Representative gating showing sorting of GLI1+ (tdTomato+) cells from Gli1-CreERT2;lox-STOP-lox-tdTomato mice.
Extended Data Fig. 6 GLI1+ cells constitute a heterogenous population.
a, Mesenchymal cells from Villin+Gli1-tdTomato mice (Villin-CreERT2;Gli1-CreERT2;lox-STOP-lox-tdTomato) 24 h after tamoxifen injection were isolated by cell sorting (see Methods). Despite the removal of the majority of epithelial cells, some epithelial cells (expressing Villin, E-cadherin and other markers) were identified as a fully distinct cluster C9. This distinct epithelial cluster serves as an internal control for scRNA-seq. Unbiased t-SNE clustering analysis of colon stem cells. Each dot represents an individual cell. 4,464 single cells were successfully profiled; numbers of cells per cluster are: C1 (n = 1,267); C2 (n = 1,072); C3 (n = 723); C4 (n = 381); C5 (n = 222); C6 (n = 159); C7 (n = 77); C8 (n = 56); C9 (n = 507). Colours correspond to unbiased classification via graph-based clustering. b, Cdh1 and Vil in epithelial cluster C9. Log-normalized gene expression levels visualized on a t-SNE plot. Each dot represents an individual cell. To show clearly mesenchymal populations, cluster C9 was removed in other panels and figures. c–f, Co-expression of indicated mesenchymal markers and Wnt ligands simultaneously visualized using t-SNE plot. Each dot represents an individual cell. Blue and red colours indicate individual genes, green colour denotes cells with simultaneously high expression of both genes. (For all panels the cluster of non-mesenchymal cells (C9) was removed.)
Extended Data Fig. 7 GLI1+ cells constitute a heterogeneous population of mesenchymal cells.
a, Expression of indicated genes in distinct populations of GLI1+ cells. Log-normalized gene expression levels visualized on a t-SNE plot. Each dot represents an individual cell. b, c, Co-expression of Myh11, Foxl1, Wnt2 and RSpo3 simultaneously visualized using t-SNE plot. Each dot represents an individual cell. Blue and red colours indicate individual genes, green colour denotes cells with simultaneous high expression of both genes. (For all panels the cluster of non-mesenchymal cells (C9) was removed; for details see Extended Data Fig. 6a.)
Extended Data Fig. 8 GLI1+ cells are enriched during epithelial perturbations.
a, Scheme of the DSS-induced damage regimen. b, Regeneration of the colonic epithelium during recovery from DSS-mediated damage is associated with an increased number of GLI1+ cells. Immunohistochemistry for GLI1 (red), DAPI (blue) and E-cadherin (green) in the colon, 3 or 5 days after the termination of DSS treatment. c, The increase in GLI1+ cells during regeneration after DSS-mediated damage does not correspond to the increased number of myeloid cells, such as macrophages (marked by F4-80) and CD11C-positive cells. Immunohistochemistry for GLI1 (red), and F4-80 (green) or CD11C (green, right) in the colon, 1 or 3 days after termination of DSS treatment. DAPI marks nuclei. Insets depict the individual green channels. The panel with CD11C staining shows a similar area to the panel with GLI1/F4-80 staining (second from right). Owing to antibody incompatibility it was not possible to perform GLI1/CD11C double staining. Scale bar, 50 μm.
Extended Data Fig. 9 In the duodenum, blocking Wnt secretion from epithelial cells is compensated by an increased number of GLI1+ cells that respond to Hedgehog pathway activation.
a, The loss of Wnt secretion in the duodenal epithelium is compensated by an increased number of GLI1+ cells in the intestinal sub-epithelial layer. Immunohistochemistry for GLI1 (red), DAPI (blue) and/or E-cadherin (green) in the duodenum. b, Quantification of GLI1+ cells in control (n = 4 independent mice) and Villin-WlscKO (n = 4 independent mice) duodena. For each mouse, three different pictures showing transverse sections were counted. The individual data points show the average number of GLI1+ cells per picture; mean ± s.d. **P ≤ 0.01, (t-test, one-sided), P = 0.008. c, Intestinal mesenchymal cells (from the duodenum) respond to stimulation of the Hedgehog pathway via recombinant SHH or smoothened agonist (SAG). Changes in the expression levels of Hedgehog target genes Gli1, Ptch1 and Ptch2 (RT–qPCR, n = 2 biologically independent experiments, each 3 replicates, mean ± s.d.). d, Activation of the Hedgehog pathway by 100 nM smoothened agonist (SAG) increased the number of GLI1+ cells in (unsorted) intestinal mesenchymal cells from the duodenum. Gli1-CreERT2;lox-STOP-lox-tdTomato+ mesenchymal cells were cultured for 5 days with (or without) 100 mM SAG and for the last 12 h with 500 nM 4-hydroxytamoxifen (4-OHT). Immunocytochemistry: tdTomato (red) marks GLI1+ cells, DAPI (blue). e, Blocking Wnt secretion from the small intestinal epithelium results in elevated levels of SHH. Immunohistochemistry: SHH (brown; DAB), nuclei (haematoxylin). f, Relative increase in the expression of Shh in the duodenum of Villin-WlscKO mice. IHH is another ligand secreted by cells of the intestinal epithelium; its expression is unchanged (RT–qPCR, 12 days after tamoxifen administration, n = 6 independent animals; mean ± s.d. *P ≤ 0.05, (one-sided t-test); P(SHH) = 0.002, P(IHH) = 0.2). Scale bar, 50 μm. Full genotypes: Villin-WlscKO (Villin-CreERT2;Wlsflox/flox).
Supplementary information
Supplementary Information
This file contains extended versions of the legends of Figs. 1–3.
Rights and permissions
About this article
Cite this article
Degirmenci, B., Valenta, T., Dimitrieva, S. et al. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453 (2018). https://doi.org/10.1038/s41586-018-0190-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0190-3
This article is cited by
-
Identifying Spatial Co-occurrence in Healthy and InflAmed tissues (ISCHIA)
Molecular Systems Biology (2024)
-
NAD+ dependent UPRmt activation underlies intestinal aging caused by mitochondrial DNA mutations
Nature Communications (2024)
-
Tumor endothelial cell-derived Sfrp1 supports the maintenance of cancer stem cells via Wnt signaling
In Vitro Cellular & Developmental Biology - Animal (2024)
-
Extracellular matrix-induced signaling pathways in mesenchymal stem/stromal cells
Cell Communication and Signaling (2023)
-
The cyclooxygenase-expressing mesenchyme resists intestinal epithelial injury by paracrine signaling
Cell Regeneration (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.