Abstract
Auxin influences plant development through several distinct concentration-dependent effects1. In the Arabidopsis root tip, polar auxin transport by PIN-FORMED (PIN) proteins creates a local auxin accumulation that is required for the maintenance of the stem-cell niche2,3,4. Proximally, stem-cell daughter cells divide repeatedly before they eventually differentiate. This developmental gradient is accompanied by a gradual decrease in auxin levels as cells divide, and subsequently by a gradual increase as the cells differentiate5,6. However, the timing of differentiation is not uniform across cell files. For instance, developing protophloem sieve elements (PPSEs) differentiate as neighbouring cells still divide. Here we show that PPSE differentiation involves local steepening of the post-meristematic auxin gradient. BREVIS RADIX (BRX) and PROTEIN KINASE ASSOCIATED WITH BRX (PAX) are interacting plasma-membrane-associated, polarly localized proteins that co-localize with PIN proteins at the rootward end of developing PPSEs. Both brx and pax mutants display impaired PPSE differentiation. Similar to other AGC-family kinases, PAX activates PIN-mediated auxin efflux, whereas BRX strongly dampens this stimulation. Efficient BRX plasma-membrane localization depends on PAX, but auxin negatively regulates BRX plasma-membrane association and promotes PAX activity. Thus, our data support a model in which BRX and PAX are elements of a molecular rheostat that modulates auxin flux through developing PPSEs, thereby timing PPSE differentiation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Benjamins, R. & Scheres, B. Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59, 443–465 (2008).
Blilou, I. et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 (2005).
Grieneisen, V. A., Xu, J., Marée, A. F., Hogeweg, P. & Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013 (2007).
Sabatini, S. et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472 (1999).
Brunoud, G. et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106 (2012).
Santuari, L. et al. Positional information by differential endocytosis splits auxin response to drive Arabidopsis root meristem growth. Curr. Biol. 21, 1918–1923 (2011).
Sauer, M. et al. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20, 2902–2911 (2006).
Geldner, N., Friml, J., Stierhof, Y. D., Jürgens, G. & Palme, K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428 (2001).
Paciorek, T. et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256 (2005).
Barbosa, I. C., Zourelidou, M., Willige, B. C., Weller, B. & Schwechheimer, C. D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev. Cell 29, 674–685 (2014).
Weller, B. et al. Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc. Natl Acad. Sci. USA 114, E887–E896 (2017).
Zourelidou, M. et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3, e02860 (2014).
Rodriguez-Villalon, A. et al. Molecular genetic framework for protophloem formation. Proc. Natl Acad. Sci. USA 111, 11551–11556 (2014).
Furuta, K. M. et al. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science 345, 933–937 (2014).
Ckurshumova, W., Smirnova, T., Marcos, D., Zayed, Y. & Berleth, T. Irrepressible MONOPTEROS/ARF5 promotes de novo shoot formation. New Phytol. 204, 556–566 (2014).
Rodriguez-Villalon, A., Gujas, B., van Wijk, R., Munnik, T. & Hardtke, C. S. Primary root protophloem differentiation requires balanced phosphatidylinositol-4,5-biphosphate levels and systemically affects root branching. Development 142, 1437–1446 (2015).
Truernit, E., Bauby, H., Belcram, K., Barthélémy, J. & Palauqui, J. C. OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 139, 1306–1315 (2012).
Scacchi, E. et al. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136, 2059–2067 (2009).
Breda, A. S., Hazak, O. & Hardtke, C. S. Phosphosite charge rather than shootward localization determines OCTOPUS activity in root protophloem. Proc. Natl Acad. Sci. USA 114, E5721–E5730 (2017).
Mouchel, C. F., Osmont, K. S. & Hardtke, C. S. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461 (2006).
Kondo, Y. et al. Vascular cell induction culture system using Arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element-like cells. Plant Cell 28, 1250–1262 (2016).
Galván-Ampudia, C. S. & Offringa, R. Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 12, 541–547 (2007).
Willige, B. C. et al. D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25, 1674–1688 (2013).
Barbosa, I. C. et al. Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143, 4687–4700 (2016).
Benková, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003).
Zádníková, P. et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137, 607–617 (2010).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153 (2003).
Bennett, T. et al. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16, 553–563 (2006).
Marchant, A. et al. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14, 589–597 (2002).
Fastner, A., Absmanner, B. & Hammes, U. Z. Use of Xenopus laevis oocytes to study auxin transport. Methods Mol. Biol. 1497, 259–270 (2017).
Absmanner, B., Stadler, R. & Hammes, U. Z. Phloem development in nematode-induced feeding sites: the implications of auxin and cytokinin. Front. Plant Sci. 4, 241 (2013).
Acknowledgements
We thank the University of Lausanne Protein Analysis Facility for mass spectrometry services, the Swiss National Science Foundation for Grant 31003A_166394 (C.S.H.), the German-Israeli Foundation for I-236-203.17-2014 (C.S.) and the Deutsche Forschungsgemeinschaft for SCHW751/12-2 (C.S.), HA 3468/6-1 and SFB924 (U.Z.H.).
Reviewer information
Nature thanks A. P. Mahonen, D. Weijers and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
P.M., A.E.L.B., M.Z., M.K., B.M., A.F., W.X.S. and P.C. performed experiments and analysed data. P.M., A.E.L.B., M.Z., W.X.S., U.Z.H., C.S. and C.S.H. designed experiments. P.M., A.E.L.B., U.Z.H., C.S. and C.S.H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Overview of protophloem development.
a, Illustration of protophloem development from the stem cell to the mature sieve element in the Arabidopsis root meristem. b, Illustration of a cross section through the stele of an Arabidopsis root meristem, highlighting the arrangement of the two sieve element strands and the xylem axis.
Extended Data Fig. 2 Auxin activity in developing PPSEs.
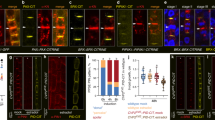
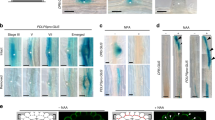
a, Confocal microscopy of the inverse auxin activity reporter DII-VENUS and its negative control mDII-VENUS (yellow fluorescence) in the root meristem (PI staining, red) of wild-type Col-0 plants. Asterisks indicate sieve element cell files. b, Confocal microscopy of constitutively expressed DII-VENUS in developing PPSEs and neighbouring cell files. Left, PI cell-wall staining (red); middle, DII-VENUS fluorescence (yellow; PPSE nuclei marked with red circles, nuclei in neighbouring cell files with blue circles); right, overlay. c, As in b, for mDII-VENUS. d, Relative intensity of the DII-VENUS reporter and its mDII-VENUS control in the nuclei of Col-0 PPSEs as compared to the nuclei of directly neighbouring cells. The statistically significant difference between DII-VENUS and mDII-VENUS in the PPSE/neighbours group is indicated (two-sided Student’s t-test; a, P = 5.86 × 10−11). e, Cumulative average cell length in different root cell files, starting from the respective first stem-cell daughters (cell #1) (n = 11 wild-type Col-0 roots). f, Number of developing PPSEs from the first stem-cell daughter up to the first transition zone PPSE (protophloem length) in seven-day-old Col-0 seedlings, and transgenic seedlings expressing a constitutively active derivative of the auxin response factor MONOPTEROS (MP∆) under control of the PPSE-specific CVP2 promoter. a, P = 3.16 × 10−6; two-sided Student’s t-test. g, Cumulative average cell length in the developing protophloem, starting from the first stem-cell daughter (cell #1) (n = 23 each). Elongation occurs prematurely in CVP2::MP∆ plants. h, Confocal microscopy of a brx root meristem, focused on one of the sieve element strands (asterisk). Arrowheads point out gap cells, which fail to build up the characteristic PPSE cell wall owing to a failure to differentiate. i, Relative intensity of the DII-VENUS reporter and its mDII-VENUS control in the nuclei of Col-0 and brx PPSEs as compared to nuclei of cells in directly neighbouring files. Statistically significant differences between PPSE/neighbours and neighbour/neighbour in the Col-0 and brx DII-VENUS groups are indicated (two-sided Student’s t-test; a, P = 2.49 × 10−7; b, P = 0.026). j, Coefficient of variance for fluorescence traces of the DII-VENUS reporter and its mDII-VENUS control (left) and PI staining (right) along protophloem cell files. The statistically significant difference in VENUS fluorescence in the brx group is indicated (two-sided Student’s t-test; a, P = 2.30 × 10−7). k, Quantification of PPSE strands with gaps in roots of indicated genotypes. l, Root length in seven-day-old seedlings for indicated genotypes. The statistically significant differences between CVP2::MPΔ in brx and brx alone (P = 0.0017) and between CVP2::MPΔ in brx and CLE45::MPΔ in brx (P = 0.0052) are indicated by the character a. m, Distribution of gap size in protophloem strands of seven-day-old seedlings with gaps of indicated genotypes. The statistically significant differences between CVP2::MPΔ in brx and brx alone (P = 0.0008) and between CVP2::MPΔ in brx and CLE45::MPΔ in brx (P = 0.0051) are indicated by the character a (two-sided χ2 test). n, Expression of fluorescent NLS–VENUS reporter in PPSEs of brx mutants, driven by either CVP2 or CLE45 promoter. Arrowheads indicate gap cells. o, p, Expression of CVP2::NLS–VENUS reporter (green fluorescence) in PPSE cell files (asterisks) of six-day-old Col-0 root meristems (PI staining, white) grown in the presence of (o), or transferred for 48 h onto (p), increasing amounts of the auxin biosynthesis inhibitor l-kynurenine (l-kyn). On the higher concentration, PPSE cell files (magnified) were barely distinguishable. q, Confocal microscopy of seven-day-old root meristems (PI staining, red). Asterisks indicate sieve element cell files (magnified, barely distinguishable in aux1 brx).
Extended Data Fig. 3 Identification of BRX interactors.
a, Induction of BRX expression in cotyledons in the VISUAL transdifferentiation assay, as indicated by a BRX::GUS reporter gene. b, Visualization of successful tracheary element differentiation using polarized light microscopy. c, Western analysis of BRX–CITRINE fusion protein after immunoprecipitation. d, List of the top BRX interactors, indicating the number of peptides isolated as compared to controls.
Extended Data Fig. 4 Phenotypic analysis of pax-related mutants and transgenic lines.
a, Root length in seven-day-old seedlings for indicated mutants and parallel Col-0 controls. Statistically significant differences between Col-0 and mutants are indicated (Student’s t-test, two-sided; a, P < 0.02). b, Quantification of gap-cell frequency in protophloem strands of six-day-old seedlings. Statistically significant differences are indicated (two-sided Fisher’s exact test; a, pax versus Col-0; b, others versus pax; all P values < 0.001). c, Root length in seven-day-old seedlings for Col-0, pax and transgenic lines in the pax mutant background that expressed PAX under the control of its native promoter or the BRX promoter. The statistically significant difference between pax and Col-0 is indicated (two-sided Student’s t-test; a, P = 0.00016). d, Quantification of gap-cell frequency in protophloem strands of six-day-old seedlings. Statistically significant differences are indicated (two-sided Fisher’s exact test; a, pax versus Col-0; b, others versus pax; all P values < 0.001). e, Phloem-mediated translocation of carboxyfluorescein diacetate succinimidyl ester (CFDA) dye (green fluorescence) into the phloem-unloading zone of the root tip 45 min after CFDA application to the cotyledons of four-day-old seedlings, and corresponding classification of CFDA signal at the end of the experiment. f, Quantification of gap-cell frequency in protophloem strands of six-day-old seedlings. Statistically significant differences are indicated (two-sided Fisher’s exact test; a, others versus Col-0; b, Col-0 and pax versus brx; all P values < 0.01). g, Quantification of gap-cell frequency in protophloem strands of six-day-old seedlings. Statistically significant differences are indicated (two-sided Fisher’s exact test; a, others versus Col-0, all P values < 0.001). h, Auxin transport assays performed in X. laevis oocytes expressing the indicated heterologous plant proteins (n = 10 oocytes per time point; error bars, s.e.m.). i, BFA control experiments. Accumulation of PIN1–GFP fusion protein in BFA compartments (left), and comparative BFA insensitivity of OPS–GFP fusion protein (right). j, Dissociation of PAX–CITRINE and BRX–CITRINE fusion proteins from the plasma membrane in response to 5 μM BFA treatment. k, Quantification of PAX–CITRINE and BRX–CITRINE fluorescence signal at the plasma membrane in response to 5 μM BFA treatment, normalized to allow direct comparison (means of approximately ten cells per root). l, Confocal microscopy of six-day-old PI-stained root meristems grown on mock or low BFA concentration as indicated. Asterisks indicate PPSE cell files and arrowheads indicate gap cells. m, Quantification of gap-cell frequency in PPSE strands of roots shown in (l). Statistically significant differences are indicated (two-sided Fisher’s exact test; a, others versus mock, P < 0.0001). n, Expression of PAX–CITRINE fusion protein under its native promoter, in pax single or brx pax double mutants. o, Transient expression of the indicated fusion proteins, alone or in combination, in Arabidopsis protoplasts. The PAXK/R>A variant carries point mutations in a polybasic stretch that is required for plasma membrane interaction24. The average number of patches per protoplast is indicated.
Extended Data Fig. 5 PIN activity in the root protophloem.
a, Confocal microscopy of indicated reporter genes (green fluorescence) in the root meristem (PI staining, red) of Col-0 wild-type plants (top), and magnification without PI background (bottom). Asterisks indicate sieve element cell files. b, Immunolocalization of nuclear localized NLS–VENUS (green) expressed under control of PPSE-specific CVP2 promoter, and PIN1 (red) by antibody staining. Asterisks indicate PPSE cell files. c, Simultaneous immunolocalization of CVP2-driven NLS–VENUS (green) with different anti-PIN1 antibodies that specifically detect phosphorylated PIN1 residues S231 (J231) or S271 (J271). d, Quantification of the J231 phosphosite signal intensity (means from approximately ten cells per root, arbitrary units). The statistically significant difference is indicated (two-sided Student’s t-test; a, P = 1.2 × 10−9). e, Quantification of the J271 phosphosite signal intensity (means from approximately ten cells per root, arbitrary units). f, Immuno-localization of PIN1, and the J231 and J271 PIN1 phosphosites (red) in brx (left) or ops (right) by antibody staining, with an OPS::BRX–CITRINE or CVP2::NLS–VENUS reporter in the background for the identification of PPSE cell files (asterisks). g, Quantification of gap-cell frequency in protophloem strands of six-day-old seedlings for the indicated genotypes. Statistically significant differences are indicated (two-sided Fisher’s exact test; a, Col-0 and pin1 versus others, P < 0.0001; b, brx or pax single mutant versus brx pin1 or pax pin1 double mutants, P < 0.02). h, Confocal microscopy of representative six-day-old Col-0, pin1, and brx root meristems (PI staining, white). Asterisks indicate PPSE cell files. i, Different phenotypic classes occurring in brx pin1 double mutant root meristems (PI staining, white). PPSE cell files were frequently barely distinguishable or missing. j, Auxin transport assays performed in X. laevis oocytes expressing the indicated heterologous plant proteins (n = 10 oocytes per time point; error bars, s.e.m.). k, Western blot analysis of the oocytes used in j, demonstrating that BRX expression does not interfere with D6PK or PIN1 expression or stability (detection of YFP–D6PK and PIN1 with anti-GFP and anti-PIN1 antibodies, respectively).
Extended Data Fig. 6 BRX auxin response and PAX specificity.
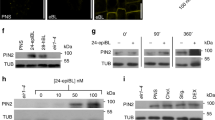
a, b, Response of BRX–CITRINE fusion protein to treatment with 1 μM or 10 μM auxin (NAA), time course experiment (b) with quantification (a, means from approximately ten cells per root, arbitrary units). Statistically significant differences are indicated (two-sided Student’s t-test; a, mock versus others, P < 0.0094; b, 1 μM versus 10 μM auxin, P < 0.0028). c, Phosphoproteomics of auxin-treated seedlings, showing normalized abundance of a conserved phosphosite in PAX, D6PK, D6PKL1-3, and AGC1-6, with subfragments indicated in different colours. d, Same as c, for a PIN1 phosphosite. e, Radioactive in vitro kinase assays with GST fusion proteins of D6PK, PAX, or PAX(S596A) and PAX(S596D) point mutants, with BRX or the PIN1 cytosolic loop as substrate (top) and corresponding loading controls (bottom). f, Auxin transport assays performed in X. laevis oocytes expressing the indicated heterologous plant proteins (n = 10 oocytes per time point; error bars, s.e.m.). g, Polar localization of the YFP–PAX(S596D) variant in developing PPSEs of a pax mutant. h, Quantification of gap-cell frequency in protophloem strands of seven-day-old pax mutant seedlings that express a D6PK–CITRINE fusion protein under the control of the PAX promoter. The statistically significant difference is indicated (two-sided Fisher’s exact test; a, Col-0 versus pax and transgenic lines, P < 0.0001). i, Polar localization of D6PK–CITRINE fusion protein in developing PPSEs of a pax mutant. j, k, Auxin induction of BRX transcription in developing PPSE cell files (asterisks) visualized using an NLS–3×VENUS reporter gene (j), with corresponding quantification of nuclear fluorescence signal (k). Statistically significant differences are indicated (one-sided Student’s t-test; a, versus preceding time point, P < 0.0153).
Extended Data Fig. 7 Molecular rheostat model for PAX–BRX action in the regulation of auxin efflux.
Proposed model for the cellular action of PAX and BRX as elements of a molecular rheostat. BRX interacts with PAX at the plasma membrane, where it inhibits PIN-mediated auxin efflux at lower auxin levels. Because of reduced PIN-mediated auxin efflux, cellular auxin levels increase so that, eventually, BRX becomes displaced from the plasma membrane. Concomitantly, PAX becomes activated and increasingly stimulates auxin efflux. Reinforced through auxin-induced BRX transcription and decreasing cellular auxin levels, BRX can return to the plasma membrane and again inhibit auxin efflux. This interplay would lead to a dynamic steady-state equilibrium that fine-tunes auxin levels along a cell file.
Supplementary information
Supplementary Information
This file contains western blot gel data.
Rights and permissions
About this article
Cite this article
Marhava, P., Bassukas, A.E.L., Zourelidou, M. et al. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558, 297–300 (2018). https://doi.org/10.1038/s41586-018-0186-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0186-z
This article is cited by
-
D6PK plasma membrane polarity requires a repeated CXX(X)P motif and PDK1-dependent phosphorylation
Nature Plants (2024)
-
Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium
Nature Plants (2023)
-
OBERON3 and SUPPRESSOR OF MAX2 1-LIKE proteins form a regulatory module driving phloem development
Nature Communications (2023)
-
A phosphoinositide hub connects CLE peptide signaling and polar auxin efflux regulation
Nature Communications (2023)
-
Ethylene response factor ERF022 is involved in regulating Arabidopsis root growth
Plant Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.