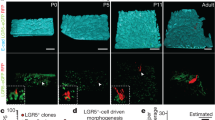
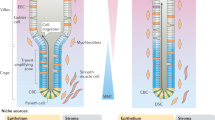
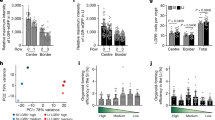
Abstract
Tissues that undergo rapid cellular turnover, such as the mammalian haematopoietic system or the intestinal epithelium, are dependent on stem and progenitor cells that proliferate to provide differentiated cells to maintain organismal health. Stem and progenitor cells, in turn, are thought to rely on signals and growth factors provided by local niche cells to support their function and self-renewal. Several cell types have been hypothesized to provide the signals required for the proliferation and differentiation of the intestinal stem cells in intestinal crypts1,2,3,4,5,6. Here we identify subepithelial telocytes as an important source of Wnt proteins, without which intestinal stem cells cannot proliferate and support epithelial renewal. Telocytes are large but rare mesenchymal cells that are marked by expression of FOXL1 and form a subepithelial plexus that extends from the stomach to the colon. While supporting the entire epithelium, FOXL1+ telocytes compartmentalize the production of Wnt ligands and inhibitors to enable localized pathway activation. Conditional genetic ablation of porcupine (Porcn), which is required for functional maturation of all Wnt proteins, in mouse FOXL1+ telocytes causes rapid cessation of Wnt signalling to intestinal crypts, followed by loss of proliferation of stem and transit amplifying cells and impaired epithelial renewal. Thus, FOXL1+ telocytes are an important source of niche signals to intestinal stem cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
05 July 2018
Change history: In this Letter, the surname of author Efi E. Massasa was misspelled ‘Massassa’. This error has been corrected online.
References
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Durand, A. et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl Acad. Sci. USA 109, 8965–8970 (2012).
Kim, T. H., Escudero, S. & Shivdasani, R. A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl Acad. Sci. USA 109, 3932–3937 (2012).
Kabiri, Z. et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 141, 2206–2215 (2014).
San Roman, A. K., Jayewickreme, C. D., Murtaugh, L. C. & Shivdasani, R. A. Wnt secretion from epithelial cells and subepithelial myofibroblasts is not required in the mouse intestinal stem cell niche in vivo. Stem Cell Reports 2, 127–134 (2014).
Aoki, R. et al. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol. Gastroenterol. Hepatol. 2, 175–188 (2016).
Kaestner, K. H. et al. Six members of the mouse forkhead gene family are developmentally regulated. Proc. Natl Acad. Sci. USA 90, 7628–7631 (1993).
Fukuda, K. et al. Mesenchymal expression of Foxl1, a winged helix transcriptional factor, regulates generation and maintenance of gut-associated lymphoid organs. Dev. Biol. 255, 278–289 (2003).
Kaestner, K. H. et al. Clustered arrangement of winged helix genes Fkh-6 and MFH-1: possible implications for mesoderm development. Development 122, 1751–1758 (1996).
Sackett, S. D., Fulmer, J. T., Friedman, J. R. & Kaestner, K. H. Foxl1-cre BAC transgenic mice: a new tool for gene ablation in the gastrointestinal mesenchyme. Genesis 45, 518–522 (2007).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Cantarero Carmona, I., Luesma Bartolomé, M. J. & Junquera Escribano, C. Identification of telocytes in the lamina propria of rat duodenum: transmission electron microscopy. J. Cell. Mol. Med. 15, 26–30 (2011).
Cretoiu, D., Cretoiu, S. M., Simionescu, A. A. & Popescu, L. M. Telocytes, a distinct type of cell among the stromal cells present in the lamina propria of jejunum. Histol. Histopathol. 27, 1067–1078 (2012).
Vannucchi, M. G., Traini, C., Manetti, M., Ibba-Manneschi, L. & Faussone-Pellegrini, M. S. Telocytes express PDGFRα in the human gastrointestinal tract. J. Cell. Mol. Med. 17, 1099–1108 (2013).
Liu, W. et al. Deletion of Porcn in mice leads to multiple developmental defects and models human focal dermal hypoplasia (Goltz syndrome). PLoS One 7, e32331 (2012).
Gracz, A. D. et al. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells 31, 2024–2030 (2013).
van der Flier, L. G., Haegebarth, A., Stange, D. E., van de Wetering, M. & Clevers, H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17 (2009).
Yan, K. S. et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+intestinal stem-cell self-renewal. Nature 545, 238–242 (2017).
Wen, F. et al. Expression of conditional Cre recombinase in epithelial tissues of transgenic mice. Genesis 35, 100–106 (2003).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Grant, G. R. et al. Comparative analysis of RNA-seq alignment algorithms and the RNA-seq unified mapper (RUM). Bioinformatics 27, 2518–2528 (2011).
Sheaffer, K. L. et al. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 28, 652–664 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Hennig, C. Cluster-wise assessment of cluster stability. Comput. Stat. Data Anal. 52, 258–271 (2007).
McLean, I. W. & Nakane, P. K. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J. Histochem. Cytochem. 22, 1077–1083 (1974).
Shyer, A. E., Huycke, T. R., Lee, C., Mahadevan, L. & Tabin, C. J. Bending gradients: how the intestinal stem cell gets its home. Cell 161, 569–580 (2015).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Itzkovitz, S. & van Oudenaarden, A. Validating transcripts with probes and imaging technology. Nat. Methods 8 (Suppl.), S12–S19 (2011).
Lyubimova, A. et al. Single-molecule mRNA detection and counting in mammalian tissue. Nat. Protoc. 8, 1743–1758 (2013).
Bahar Halpern, K. et al. Bursty gene expression in the intact mammalian liver. Mol. Cell 58, 147–156 (2015).
Bahar Halpern, K. & Itzkovitz, S. Single molecule approaches for quantifying transcription and degradation rates in intact mammalian tissues. Methods 98, 134–142 (2016).
Itzkovitz, S. et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 14, 106–114 (2011).
Acknowledgements
We thank A. Rustgi, C. Lengner and Y. Dor for critical reading of the manuscript; M. Sundaram for suggesting the porcupine gene ablation experiment; and all members of the Kaestner laboratory. We acknowledge funding from NIDDK through R37-DK053839. We thank the University of Pennsylvania’s Diabetes Research Center (DRC) for the use of the Functional Genomics Core (P30-DK019525) and the Center for Molecular Studies in Digestive and Liver Diseases for the use of the Molecular Pathology and Imaging and Transgenic and Chimeric Mouse Cores (P30-DK050306).
Reviewer information
Nature thanks L. Samuelson and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.S.-C. and K.H.K. came up with the study concept and design, analysed and interpreted the data and wrote and revised the manuscript. B.T., E.E.M. and S.I. acquired and interpreted smFISH data and reviewed the manuscript. Y.J.W., K.J.W. and A.K. acquired and interpreted immunostaining and RNA-seq data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1
Fluorescence-activated cell sorting plots. a–d, Representative FACS plots of mesenchymal cells isolated from Foxl1-cre;Rosa26-mTmG (c, d) as compared to control Rosa26-mTmG (a, b) mice, showing sorting strategy. In a and c, Allophycocyanin+ (APC+), CD45+ and EPCAM+-labelled cells were gated out to exclude immune and epithelial cell contamination. In d, FOXL1+, GFP+ and FOXL1−Tomato+ cells were gated and sorted based on fluorescence activity as compared to the negative control (b). Experiments were repeated at least three times with similar results.
Extended Data Fig. 2
Single-cell qPCR of FOXL1+ telocytes showing heterogeneity within the FOXL1+ cell population. Hierarchical clustering of FOXL1+ single cells based on qPCR-based detection of 30 genes. Note that all FOXL1+ cells express Pdgfra and Wnt3a. The population is clustered into three main groups. Jaccard coefficients for the three clusters were: 0.83 (green), 0.69 (turquoise) and 0.79 (yellow), respectively, indicating underlying cluster stability. Values between 0.6 and 0.75 indicate that the cluster is measuring a pattern in the data. Clusters with stability values above about 0.85 are considered to be highly stable.
Extended Data Fig. 3
Derivation of Foxl1-creERT2;PorcnΔ mice. a, Schema for the generation of Foxl1-creERT2 mice using BAC recombineering. The coding sequence of exon 1 of Foxl1 was targeted by the sequence of creERT2. FRT, flipase recognition target; Flp, flipase; LA, left homology arm; RA, right homology arm. b, c, Tamoxifen induction of Foxl1-creERT2;Rosa26-mTmG-driven expression of membrane-bound GFP to mesenchymal telocytes in the duodenum (b) and colon (c). Immunofluorescence staining for GFP (green), EPCAM (red). d, Schema for the generation of Foxl1-creERT2;PorcnΔ mice. Foxl1-creERT2 mice were crossed with mice carrying loxP sites flanking exons 3–7 of the X-linked porcupine homologue (Porcn) gene. e, Foxl1-creERT2;Rosa26-mTmG (control, n = 5 mice) and Foxl1-creERT2;PorcnΔ (PorcnΔ, n = 8 mice) male mice were treated with tamoxifen for three consecutive days to induce Cre expression and weighed every day. The slope of the weight loss was significantly different in PorcnΔ mice as compared to control mice (*P = 0.0107, two-tailed linear regression analysis).
Supplementary information
Video 1:
Foxl1+ telocytes form a continuous subepithelial network from crypt base to villus tips. Confocal 3D projection of cleared duodenum from Foxl1Cre; Rosa-mTmG mouse stained with GFP (green), EpCAM (red) and DAPI. Image width is 1.2 mm. The stained intestine was placed en block on an image slide using 1 mm depth adhesive silicone isolator, mounted in X-CLARITYTM mounting solution and imaged using confocal scanning. Z-stacks projections were compiled using Volocity software
Rights and permissions
About this article
Cite this article
Shoshkes-Carmel, M., Wang, Y.J., Wangensteen, K.J. et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246 (2018). https://doi.org/10.1038/s41586-018-0084-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0084-4
This article is cited by
-
NAD+ dependent UPRmt activation underlies intestinal aging caused by mitochondrial DNA mutations
Nature Communications (2024)
-
Intestinal stem cells: guardians of homeostasis in health and aging amid environmental challenges
Experimental & Molecular Medicine (2024)
-
Dynamic relationship among extracellular matrix and body wall cells in Hirudo verbana morphogenesis
Cell and Tissue Research (2024)
-
Telocytes: current methods of research, challenges and future perspectives
Cell and Tissue Research (2024)
-
A stromal lineage maintains crypt structure and villus homeostasis in the intestinal stem cell niche
BMC Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.