Abstract
Ribosomal surveillance pathways scan for ribosomes that are transiently paused or terminally stalled owing to structural elements in mRNAs or nascent chain sequences1, 2. Some stalls in budding yeast are sensed by the GTPase Hbs1, which loads Dom34, a catalytically inactive member of the archaeo-eukaryotic release factor 1 superfamily. Hbs1–Dom34 and the ATPase Rli1 dissociate stalled ribosomes into 40S and 60S subunits. However, the 60S subunits retain the peptidyl-tRNA nascent chains, which recruit the ribosome quality control complex that consists of Rqc1–Rqc2–Ltn1–Cdc48–Ufd1–Npl4. Nascent chains ubiquitylated by the E3 ubiquitin ligase Ltn1 are extracted from the 60S subunit by the ATPase Cdc48–Ufd1–Npl4 and presented to the 26S proteasome for degradation3,4,5,6,7,8,9. Failure to degrade the nascent chains leads to protein aggregation and proteotoxic stress in yeast and neurodegeneration in mice10,11,12,13,14. Despite intensive investigations on the ribosome quality control pathway, it is not known how the tRNA is hydrolysed from the ubiquitylated nascent chain before its degradation. Here we show that the Cdc48 adaptor Vms1 is a peptidyl-tRNA hydrolase. Similar to classical eukaryotic release factor 1, Vms1 activity is dependent on a conserved catalytic glutamine. Evolutionary analysis indicates that yeast Vms1 is the founding member of a clade of eukaryotic release factor 1 homologues that we designate the Vms1-like release factor 1 clade.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Buskirk, A. R. & Green, R. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Phil. Trans. R. Soc. Lond. B 372, 20160183 (2017).
Joazeiro, C. A. P. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol. 33, 343–368 (2017).
Ito-Harashima, S., Kuroha, K., Tatematsu, T. & Inada, T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21, 519–524 (2007).
Bengtson, M. H. & Joazeiro, C. A. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010).
Shoemaker, C. J. & Green, R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl Acad. Sci. USA 108, E1392–E1398 (2011).
Brandman, O. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012).
Verma, R., Oania, R. S., Kolawa, N. J. & Deshaies, R. J. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife 2, e00308 (2013).
Defenouillère, Q. et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl Acad. Sci. USA 110, 5046–5051 (2013).
Shao, S., von der Malsburg, K. & Hegde, R. S. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648 (2013).
Shen, P. S. et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 (2015).
Chu, J. et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl Acad. Sci. USA 106, 2097–2103 (2009).
Choe, Y. J. et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 (2016).
Yonashiro, R. et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife 5, e11794 (2016).
Defenouillère, Q. et al. Rqc1 and Ltn1 prevent C-terminal alanine-threonine tail (CAT-tail)-induced protein aggregation by efficient recruitment of Cdc48 on stalled 60S subunits. J. Biol. Chem. 291, 12245–12253 (2016).
Blythe, E. E., Olson, K. C., Chau, V. & Deshaies, R. J. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP•NPLOC4•UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc. Natl Acad. Sci. USA 114, E4380–E4388 (2017).
Bodnar, N. O. & Rapoport, T. A. Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722–735.e9 (2017).
Hänzelmann, P. & Schindelin, H. The structural and functional basis of the p97/valosin-containing protein (VCP)-interacting motif (VIM): mutually exclusive binding of cofactors to the N-terminal domain of p97. J. Biol. Chem. 286, 38679–38690 (2011).
Stapf, C., Cartwright, E., Bycroft, M., Hofmann, K. & Buchberger, A. The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors. J. Biol. Chem. 286, 38670–38678 (2011).
van Hoof, A., Frischmeyer, P. A., Dietz, H. C. & Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 (2002).
Alamgir, M., Erukova, V., Jessulat, M., Azizi, A. & Golshani, A. Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chem. Biol. 10, 6 (2010).
Shao, S. & Hegde, R. S. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell 55, 880–890 (2014).
Nielson, J. R. et al. Sterol oxidation mediates stress-responsive Vms1 translocation to mitochondria. Mol. Cell 68, 673–685.e6 (2017).
Song, H. et al. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321 (2000).
Jin, H., Kelley, A. C., Loakes, D. & Ramakrishnan, V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc. Natl Acad. Sci. USA 107, 8593–8598 (2010).
Cheng, Z. et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 23, 1106–1118 (2009).
Korostelev, A. et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl Acad. Sci. USA 105, 19684–19689 (2008).
Greber, B. J. et al. Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell 164, 91–102 (2016).
Preis, A. et al. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 8, 59–65 (2014).
Izawa, T., Park, S. H., Zhao, L., Hartl, F. U. & Neupert, W. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell 171, 890–903.e18 (2017).
von der Malsburg, K., Shao, S. & Hegde, R. S. The ribosome quality control pathway can access nascent polypeptides stalled at the Sec61 translocon. Mol. Biol. Cell 26, 2168–2180 (2015).
Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 (2001).
Pierce, N. W. et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153, 206–215 (2013).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics 10, M111.013284 (2011).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Eddy, S. R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23, 205–211 (2009).
Alva, V., Nam, S. Z., Söding, J. & Lupas, A. N. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 44, W410–W415 (2016).
Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008).
Lassmann, T., Frings, O. & Sonnhammer, E. L. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865 (2009).
Cole, C., Barber, J. D. & Barton, G. J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 (2008).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Acknowledgements
We thank A. Buchberger, H. Rao, A. van Hoof, R. Voorhees and J. Warner for reagents; the Proteome Exploration Laboratory (PEL), Caltech, for help with the mass spectrometry analysis and the members of the Deshaies laboratory, M. Blanco, M. Guttman, B. Clemons, R. Voorhees and S. Shan for discussions. R.J.D. was an investigator of the HHMI and this work was funded in part by the HHMI. A.M.B. and L.A. are supported by the funds of the Intramural Research Program of the National Library of Medicine.
Author information
Authors and Affiliations
Contributions
R.V., K.M.R. and R.J.D. designed experiments, J.M.R. performed mass spectrometry analyses, L.A. and A.M.B. did the computational analyses, K.M.R. performed the mutagenesis analyses, R.S.O. generated all the yeast strains and R.V. performed all the biochemical experiments. R.V. and R.J.D. supervised research, R.V., R.J.D. and L.A. wrote the paper and all authors participated in editing the manuscript. All figure schematics were generated by K.M.R.
Corresponding authors
Ethics declarations
Competing interests
R.V., K.M.R., A.M.B., R.S.O., J.M.R. and L.A. declare no competing interests. R.J.D. is currently Senior Vice President of discovery research at Amgen and a Visiting Associate at the California Institute of Technology (Caltech).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
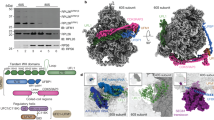
Extended Data Fig. 1 Mass spectrometry analysis of Vms1 required for release of Ub–PrANS–tRNA from ribosomes.
a, Serial tenfold dilutions of exponential cultures were spotted on YPD plates with or without 100 μg ml−1 hygromycin B and allowed to grow at 30 °C for 3 days. Data shown are representative of three biological replicates. b, Ponceau S stained nitrocellulose filter used as loading control for Fig. 1c. c, Lysate from vms1Δ cells expressing PrANS (input lysate, lane five) was fractionated on a sucrose gradient and 60S fractions were pooled and mock-treated (lanes one and three) or pretreated with the deubiquitylating enzyme Usp2c (1 μM; lane two) or 100 μg ml−1 RNase A (lane four) at 30 °C for 20 min before CTAB precipitation. The pellets were immunoblotted with PAP. d, Ribosomes from the indicated strains were isolated using sucrose cushions and aliquots were immunoblotted to detect PrANS and Rpl3. The remainder was bound to TUBE resin. The adsorbed fractions were immunoblotted with PAP. All lanes in the left and right panels are from the same blots. The dashed lines indicate cropping of lanes not pertinent to the current study. Uncropped gel source images are provided in Supplementary Fig. 1. Data in c and d are representative of two biological replicates. e–h, Mass spectrometry analysis of TAP-tagged Cdc48, Vms1 and Ufd1. e, Relative abundance of each Cdc48 adaptor co-immunoprecipitated with Cdc48 relative to all adaptor proteins identified. f, Schematic illustrating the estimated stoichiometry of Cdc48–adaptor complexes. g, Relative stoichiometry of associated proteins that co-immunoprecipitated with Cdc48, Vms1 and Ufd1. Samples were normalized to the iBAQ value of the bait protein and are presented as percentage of the bait protein. Protein iBAQ values from the untagged control were subtracted from the tagged immunoprecipitation samples. h, Coomassie blue-stained gel of samples used for mass spectrometry analysis. All mass spectrometry data (e–h) are representative of two biological replicates.
Extended Data Fig. 2 Vms1 is the founding member of the VLRF1 clade.
a, Extended sequence alignment of aeRF1 superfamily with representatives from all families and clades (compared to the limited subset shown in Fig. 2a). b, Phylogenetic tree depicting relationships within the aeRF1 superfamily; colouring matches clade labels in Fig. 2a. In the classical aeRF1 clade, two branches contain eukaryotic orthologues (eRF1) and archaeal orthologues (aRF1), respectively. Of the bacterial (baeRF1) versions, certain members are misannotated as ‘Host_attach’ in the Pfam database while most cannot be detected by existing profiles. The total number of prokaryotic representatives of the VLRF1 clade in the non-redundant database (NCBI, as of 1 December 2017) is 1,044. Of these, the archaeal VLRF1 family (aVLRF1) has 279 members, actino-chloroflexi VLRF1 family (acVLRF1) has 669 and the bacteroidetes VLRF1 family (bVLRF1) has 96. Notable domain architectures and conserved gene neighbourhoods are shown to the right of tree. A gene encoding a ribosome hibernation factor (HPF-like or YfiA-like) that facilitates inactive ribosome aggregation frequently co-occurs and is predicted to function with the baeRF1 domains. The labels below the architecture diagrams list the well-characterized clade representatives, PDB structure accessions and phyletic distributions. The dashed outline indicates domains that are not universally present.
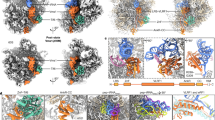
Extended Data Fig. 3 Mutagenesis of the non-catalytic domains of Vms1.
a, SDS lysates of cells were immunoblotted with anti-haemagglutinin antibodies. Ponceau S staining of the blot shows the equivalency of the extracts. The RR mutant is R313A, R314A. The DNKR mutant contains the following four mutations in the zinc-finger domain: D94A, N99A, K101A, and R102A. The ∆VIM mutant is deleted for amino acids 622–625. b, Serial tenfold dilutions of wild-type and vms1Δ cells transformed with the indicated vms1 alleles were spotted on YPD plates containing 10 or 25 ng ml−1 cycloheximide and incubated at 30 °C for 3 days. Data are representative of three biological replicates. c, Native lysates (10 A260 units each) were subjected to sucrose gradient analysis. In each case fractions 10–23 were resolved and immunoblotted to detect protein A and Rpl3. Identical exposures are shown for all panels. d, Native lysates of vms1Δ cells expressing wild-type Vms1–HA3 or Vms1–HA3-DNKR were subjected to sucrose gradient analysis. Fractions were immunoblotted for Vms1 and Rpl3. MW, molecular weight marker. All western blot data are representative of two biological replicates. Gel source images are available in Supplementary Fig. 1.
Extended Data Fig. 4 In vitro reconstitution of Vms1 peptidyl-tRNA hydrolase activity.
a, Coomassie blue-stained gels of the indicated purified proteins used for in vitro reconstitution. b, Analysis of ribosome–nascent chain complexes generated by translation in reticulocyte lysate. PCR products encoding Flag–CRPNS followed by a 30-nucleotide poly(A) sequence and Flag–CRPStop (with a stop codon) were transcribed and translated in reticulocyte lysate in the presence of 35S-labelled methionine. Completed translation reactions were treated (or not treated), fractionated using SDS–PAGE, and visualized by autoradiography. Lane one, no treatment; lane two, RNase A treatment; lane three, CTAB treatment; lane four, immunoprecipitated with anti-Flag resin. For lanes three and four, the pellet fraction was analysed. c, Sucrose gradient analysis of reticulocyte translation reactions. Flag–CRPNSKn was transcribed and translated in 200 μl reticulocyte lysate for 30 min. Lysates were layered onto 2-ml 10–50% sucrose gradients and centrifuged using a Beckman SW55 rotor for 80 min. Eleven fractions (200 μl each) were collected from the top by hand. Aliquots were analysed for fractionation of 35S-labelled substrate and Rpl8 by autoradiography and immunoblotting, respectively. d, Quantification of two independent biological replicates of the yeast His6–Vms1 and His6–Vms1-Q295L titration reactions shown in Fig. 4d. e, Quantification of the human ANKZF1 and ANKZF1-Q246L titration reactions shown in Fig. 4e. Data are representative of two biological replicates. Gel source images are provided in Supplementary Fig. 1 and Source Data are available with the online version of this paper.
Extended Data Fig. 5 Working model of Vms1 function at stalled ribosomes.
In non-stop decay of mRNAs lacking a stop codon, ribosomes translate the poly(A) tail and stall after translating several lysines4, 19. The A site as a consequence is occupied by an AAA codon. Our data suggest that Vms1 can potentially hydrolyse peptidyl-tRNA chains on this leading stalled ribosome without prior splitting by Dom34–Hbs1–Rli1. One of the known responses to stalling is endonucleolytic cleavage of the mRNA by an as-yet unidentified endonuclease. The cleavage reaction generates a truncated transcript. Lagging ribosomes that translate up to the cleavage site stall with an empty A site. Such stalls are recognized by Dom34–Hbs1 that together with Rli1 dissociate the 80S ribosome into the 40S subunit and the 60S subunit, which contains the nascent peptidyl-tRNA1. Dissociation allows for stable association of the RQC complex members Rqc1–Rqc2 and the E3 ubiquitin ligase Ltn19. Rqc2 adds non-templated alanine and threonine residues to the C-terminal end of the nascent chain to extrude sequences in the exit tunnel past the active site of Ltn1, which ubiquitylates lysine residues in the emerging nascent chain10 and with the aid of Cdc48, optimizes the conformation of Vms1 at the PTC such that it can hydrolyse the tRNA. The ubiquitylated nascent chain is engaged by Ufd1–Npl4 bound to Cdc48 that together unfold and extract the nascent chain. Dissociation of Rqc2 allows Vms1 to access the 60S subunit29, resulting in hydrolysis of the peptidyl-tRNA. Recruitment of Cdc48–Ufd1–Npl4 to the ubiquitylated nascent chain7 ensures its efficient extraction from the 60S subunit. It is unclear whether the action of Vms1 on 80S ribosomes is coupled in some manner to ribosome splitting.
Supplementary information
Supplementary Figure 1
This file contains the gel source data
Source data
Rights and permissions
About this article
Cite this article
Verma, R., Reichermeier, K.M., Burroughs, A.M. et al. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature 557, 446–451 (2018). https://doi.org/10.1038/s41586-018-0022-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0022-5
This article is cited by
-
Stalled translation by mitochondrial stress upregulates a CNOT4-ZNF598 ribosomal quality control pathway important for tissue homeostasis
Nature Communications (2024)
-
Genes controlling hydrolysate toxin tolerance identified by QTL analysis of the natural Saccharomyces cerevisiae BCC39850
Applied Microbiology and Biotechnology (2024)
-
Valosin containing protein (VCP): initiator, modifier, and potential drug target for neurodegenerative diseases
Molecular Neurodegeneration (2023)
-
Mitochondrial complexome reveals quality-control pathways of protein import
Nature (2023)
-
Nascent peptide-induced translation discontinuation in eukaryotes impacts biased amino acid usage in proteomes
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.