Abstract
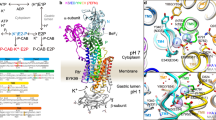
The gastric proton pump—the H+, K+-ATPase—is a P-type ATPase responsible for acidifying the gastric juice down to pH 1. This corresponds to a million-fold proton gradient across the membrane of the parietal cell, the steepest known cation gradient of any mammalian tissue. The H+, K+-ATPase is an important target for drugs that treat gastric acid-related diseases. Here we present crystal structures of the H+, K+-ATPase in complex with two blockers, vonoprazan and SCH28080, in the luminal-open state, at 2.8 Å resolution. The drugs have partially overlapping but clearly distinct binding modes in the middle of a conduit running from the gastric lumen to the cation-binding site. The crystal structures suggest that the tight configuration at the cation-binding site lowers the pK a value of Glu820 sufficiently to enable the release of a proton even into the pH 1 environment of the stomach.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
05 April 2018
The Extended Data Figures and Tables section originally published with this article was missing Tables 1–3. This has now been corrected.
References
Ganser, A. L. & Forte, J. G. K+-stimulated ATPase in purified microsomes of bullfrog oxyntic cells. Biochim. Bioshys. Acta 307, 169–180 (1973).
Sachs, G. et al. The gastric H,K ATPase as a drug target: past, present, and future. J. Clin. Gastroenterol. 41, S226–S242 (2007).
Sachs, G., Meyer-Rosberg, K., Scott, D. R. & Melchers, K. Acid, protons and Helicobacter pylori. Yale J. Biol. Med. 69, 301–316 (1996).
Otake, K. et al. Characteristics of the novel potassium-competitive acid blocker vonoprazan fumarate (TAK-438). Adv. Ther. 33, 1140–1157 (2016).
Kaminski, J. J., Wallmark, B., Briving, C. & Andersson, B. M. Antiulcer agents. 5. Inhibition of gastric H+/K+-ATPase by substituted imidazo[1,2-a]pyridines and related analogues and its implication in modeling the high affinity potassium ion binding site of the gastric proton pump enzyme. J. Med. Chem. 34, 533–541 (1991).
Shin, J. M. & Sachs, G. Pharmacology of proton pump inhibitors. Curr. Gastroenterol. Rep. 10, 528–534 (2008).
Rabon, E. C. & Reuben, M. A. The mechanism and structure of the gastric H,K-ATPase. Annu. Rev. Physiol. 52, 321–344 (1990).
Rabon, E. C., McFall, T. L. & Sachs, G. The gastric [H,K]ATPase:H+/ATP stoichiometry. J. Biol. Chem. 257, 6296–6299 (1982).
Morth, J. P. et al. Crystal structure of the sodium–potassium pump. Nature 450, 1043–1049 (2007).
Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 (2000).
Wolosin, J. M. Ion transport studies with H+-K+-ATPase-rich vesicles: implications for HCl secretion and parietal cell physiology. Am. J. Physiol. Gastrointest. Liver Physiol. 248, G595–G607 (1985).
Dukkipati, A., Park, H. H., Waghray, D., Fischer, S. & Garcia, K. C. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif. 62, 160–170 (2008).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protocols 9, 2574–2585 (2014).
Bublitz, M., Poulsen, H., Morth, J. P. & Nissen, P. In and out of the cation pumps: P-type ATPase structure revisited. Curr. Opin. Struct. Biol. 20, 431–439 (2010).
Abe, K., Tani, K. & Fujiyoshi, Y. Systematic comparison of molecular conformations of H+,K+-ATPase reveals an important contribution of the A-M2 linker for the luminal gating. J. Biol. Chem. 289, 30590–30601 (2014).
Olesen, C. et al. The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 (2007).
Laursen, M., Yatime, L., Nissen, P. & Fedosova, N. U. Crystal structure of the high-affinity Na+K+-ATPase–ouabain complex with Mg2+ bound in the cation binding site. Proc. Natl Acad. Sci. USA 110, 10958–10963 (2013).
Toyoshima, C., Norimatsu, Y., Iwasawa, S., Tsuda, T. & Ogawa, H. How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump. Proc. Natl Acad. Sci. USA 104, 19831–19836 (2007).
Abe, K., Tani, K., Nishizawa, T. & Fujiyoshi, Y. Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle. EMBO J. 28, 1637–1643 (2009).
Abe, K., Tani, K., Friedrich, T. & Fujiyoshi, Y. Cryo-EM structure of gastric H+,K+-ATPase with a single occupied cation-binding site. Proc. Natl Acad. Sci. USA 109, 18401–18406 (2012).
Abe, K., Tani, K. & Fujiyoshi, Y. Conformational rearrangement of gastric H+,K+-ATPase induced by an acid suppressant. Nat. Commun. 2, 155 (2011).
Scott, D. R., Munson, K. B., Marcus, E. A., Lambrecht, N. W. G. & Sachs, G. The binding selectivity of vonoprazan (TAK-438) to the gastric H+, K+ -ATPase. Aliment. Pharmacol. Ther. 42, 1315–1326 (2015).
Danko, S., Yamasaki, K., Daiho, T. & Suzuki, H. Distinct natures of beryllium fluoride-bound, aluminum fluoride-bound, and magnesium fluoride-bound stable analogues of an ADP-insensitive phosphoenzyme intermediate of sarcoplasmic reticulum Ca2+-ATPase: changes in catalytic and transport sites during phosphoenzyme hydrolysis. J. Biol. Chem. 279, 14991–14998 (2004).
Danko, S., Yamasaki, K., Daiho, T. & Suzuki, H. Membrane perturbation of ADP-insensitive phosphoenzyme of Ca2+-ATPase modifies gathering of transmembrane helix M2 with cytoplasmic domains and luminal gating. Sci. Rep. 7, 41172 (2017).
Jorgensen, P. L., Håkansson, K. O. & Karlish, S. J. D. Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 65, 817–849 (2003).
Koenderink, J. B., Swarts, H. G. P., Willems, P. H. G. M., Krieger, E. & De Pont, J. J. H. H. M. A conformation-specific interhelical salt bridge in the K+ binding site of gastric H,K-ATPase. J. Biol. Chem. 279, 16417–16424 (2004).
Munson, K., Garcia, R. & Sachs, G. Inhibitor and ion binding sites on the gastric H,K-ATPase. Biochemistry 44, 5267–5284 (2005).
Burnay, M., Crambert, G., Kharoubi-Hess, S., Geering, K. & Horisberger, J. D. Electrogenicity of Na,K- and H,K-ATPase activity and presence of a positively charged amino acid in the fifth transmembrane segment. J. Biol. Chem. 278, 19237–19244 (2003).
Dürr, K. L., Seuffert, I. & Friedrich, T. Deceleration of the E1P–E2P transition and ion transport by mutation of potentially salt bridge-forming residues Lys-791 and Glu-820 in gastric H+/K+-ATPase. J. Biol. Chem. 285, 39366–39379 (2010).
Sielecki, A. R., Fedorov, A. A., Boodhoo, A., Andreeva, N. S. & James, M. N. G. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 Å resolution. J. Mol. Biol. 214, 143–170 (1990).
Clement, G. E., Rooney, J., Zakheim, D. & Eastman, J. The pH dependence of the dephosphorylated pepsin-catalyzed hydrolysis of N-acetyl-l-phenylalanyl-l-tyrosine methyl ester. J. Am. Chem. Soc. 92, 186–189 (1970).
Asano, S., Furumoto, R., Tega, Y., Matsuda, S. & Takeguchi, N. Mutational analysis of the putative K+-binding site on the fourth transmembrane segment of the gastric H+,K+-ATPase. J. Biochem. 127, 993–1000 (2000).
Palmgren, M. G., Buch-Pedersen, M. J. & Møller, A. L. Mechanism of proton pumping by plant plasma membrane H+-ATPase: role of residues in transmembrane segments 5 and 6. Ann. NY Acad. Sci. 986, 188–197 (2003).
Pedersen, B. P., Buch-Pedersen, M. J., Morth, J. P., Palmgren, M. G. & Nissen, P. Crystal structure of the plasma membrane proton pump. Nature 450, 1111–1114 (2007).
Focht, D., Croll, T. I., Pedersen, B. P. & Nissen, P. Improved model of proton pump crystal structure obtained by interactive molecular dynamics flexible fitting expands the mechanistic model for proton translocation in P-type ATPases. Front. Physiol. 8, 202 (2017).
Jorgensen, P. L. & Amat, F. Regulation and function of lysine-substituted Na,K pumps in salt adaptation of Artemia franciscana. J. Membr. Biol. 221, 39–49 (2008).
Gourdon, P. et al. HiLiDe—systematic approach to membrane protein crystallization in lipid and detergent. Cryst. Growth Des. 11, 2098–2106 (2011).
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Abe, K. et al. The cryo-EM structure of gastric H+,K+-ATPase with bound BYK99, a high-affinity member of K+-competitive, imidazo[1,2-a]pyridine inhibitors. Sci. Rep. 7, 6632 (2017).
Kanai, R., Ogawa, H., Vilsen, B., Cornelius, F. & Toyoshima, C. Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state. Nature 502, 201–206 (2013).
Acknowledgements
We thank M. Taniguchi for the technical assistance; T. Imagawa for the cDNA of pig gastric H+, K+-ATPase; P. Gourdon for sharing the crystal screening matrix; D. McIntosh for improving the manuscript; P. Nissen and C. Toyoshima for critical discussion; and K. Taniguchi for his support from the initial stage of this project. This work was supported by Grants-in-Aid for Scientific Research (B), CREST from JST (JPMJCR14M4), and Basis for Supporting Innovative Drug Discovery and Life Science Research (to K.A.); Grants-in-Aid for Scientific Research (S), the Japan New Energy and Industrial Technology Development Organization (NEDO), and the Japan Agency for Medical Research and Development (AMED) (to Y.F.). The synchrotron radiation experiments were performed at BL32XU and BL41XU in SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI Proposal numbers: 2011B1240, 2014A1248, 2014B1165, 2015B1042, 2016B2721 and 2017B2701). We thank the beamline staff for their facilities and support.
Reviewer information
Nature thanks M. Palmgren, H. Poulsen and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
K.A. and Y.F. designed the study. K.A. and H.S. expressed the proteins. K.A. and H.N. purified and crystallized the proteins. K.A. performed the biochemical analysis. K.A., H.N. and K.I. corrected the X-ray diffraction data. K.A. and K.I. analysed the structures. All authors interpreted the structure and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Y.F. is a director of CeSPIA Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Crystallization of gastric H+, K+-ATPase.
a, Purification of H+, K+-ATPase expressed in HEK293 cells. Lane 1: solubilized membrane fraction, lane 2: pass through of Flag resin, lane 3: wash fraction, lane 4: elution by Flag peptide, lane 5: TEV protease- and endoglycosidase-treated sample, lane 6: pass-through fraction of Ni-NTA and amylose resin, lane 7: concentrated peak fractions by size-exclusion chromatography. b, The elution profile of affinity-purified H+, K+-ATPase by Superose6 Increase 10/300. Black, red and green arrowheads indicate elution volume of aggregation, α–β-complex of H+, K+-ATPase and cleaved EGFP, respectively. Purification was well reproduced, and representative results are shown in the figure. c, d, Crystals of H+, K+-ATPase in the presence of vonoprazan (c) or SCH28080 and Rb+ (d). Scale bars, 100 μm. e, X-ray diffraction of the vonoprazan-bound crystal. Enlarged image shows diffraction spots of up to 2.3 Å in the direction of the c* axis, although the crystal shows anisotropic diffractions. Most crystals showed diffraction spots of up to 2.8 Å in similar crystallization conditions and a few crystals showed diffraction spots better than 2.3 Å, as shown in the figure. f, Crystal packing. An asymmetric unit (molecule in the lower left, depicted as in Fig. 1d) contains one α–β-complex of H+, K+-ATPase (α-subunit, light blue; β-subunit, wheat; vonoprazan, magenta), packed with P3121 symmetry. A unit cell and approximate location of the membrane planes are provided as grey and yellow boxes, respectively.
Extended Data Fig. 2 Crystal structure of gastric H+, K+-ATPase bound to SCH28080.
a, Overall structure of the luminal-open E2P state of H+, K+-ATPase complexed with SCH28080 ((SCH)E2BeF) in the ribbon representations, as in Fig. 1a. Bound SCH28080 and three Rb+ ions are shown as green and purple spheres, respectively. Inset, chemical structure of SCH28080. b, Magenta mesh shows anomalous peaks from Rb+ contoured at the 5σ level, indicating that three Rb+ ions (blue, yellow and red boxes) are bound to the H+, K+-ATPase (SCH)E2BeF (shown as colour ribbons). Blue, interface between the nucleotide domain and the actuator domain of the symmetry-related neighbouring molecules (grey ribbon). Yellow, K+-binding site at the phosphorylation domain, which is homologous to SERCA and Na+, K+-ATPase. Red, anomalous peak found at the transmembrane cation-binding site. c, The molecular surface of (SCH)E2BeF structure, viewed from the luminal side of the membrane. Bound SCH28080 (green sticks) blocks the conduit connecting to the cation-binding site. d, Structure as in c, but with bound SCH28080 is removed, showing that Rb+ bound to the cation-binding site (purple) is exposed to the luminal solution. e–g, The Cα traces of the indicated atomic models are superimposed on the H+, K+-ATPase (Von)E2BeF (blue, with bound vonoprazan shown as spheres).
Extended Data Fig. 3 TM2 helix and the hydrophobic cluster.
a–d, Interface between the actuator and phosphorylation domains, and the cytoplasmic portion of the TM2 in the luminal-open E2P state of H+, K+-ATPase (Von)E2BeF (a, c) and the luminal-closed E2P transition state of SERCA E2-AlF (PDB code: 2ZBG)18 (b, d) are shown. These two atomic models are superimposed according to the TM7–TM10 structure. Broken box on the whole molecular structure (upper left) indicates the region shown in a–d, viewed from left (a, b) or front (c, d) of the molecule. Actuator domain (green), TM1–TM2 (blue), and TM3–TM4 (cyan) bundles are highlighted. Residues that contribute to the hydrophobic interactions19 (orange dotted circles) are indicated as spheres with analogous colouring of their respective structural components. Phe170 in H+, K+-ATPase is homologous to Tyr122 in SERCA. Because of the different coordination geometry between phosphate analogues (BeF3 −, light blue; AlF4 −, pink) and the TGES motif (indicated as dark colour in each model) at the interface between the actuator and phosphorylation domains (see Fig. 1e for closed view), the azimuthal position of the actuator domain differs between the two structures (by approximately 30°, as indicated by the orange arrow in b). As a consequence, the cytoplasmic portion of TM2 shows different conformations between the α-helical structure in the luminal-open E2P (a, c) and unwound loop structure in the luminal-closed E2-P forms (b, d). The Cα positions of Ile119 and Met334 in H+, K+-ATPase (a gating latch) and their homologous residues in SERCA (Ile71 and Val300) are shown in red (see Fig. 3). e, Schematic of luminal gate closure in H+, K+-ATPase. In the luminal-open E2P state (left), Ile119 (TM1) and Met334 (TM4) act as a latch to keep the TM1–TM2 bundle in the upright cytoplasmic-side position (indicated by dotted lines and arrows). Binding of counter-transporting K+ to the cation-binding site induces luminal gate closure (right), which is accompanied by the lateral movement of the TM3–TM4 bundle (Fig. 1c) and downward-sliding movement of the TM1–TM2 bundle (indicated by red arrows in the left panel). The sliding movement of TM1–TM2 results in the unwinding of the cytoplasmic portion of TM2 and the rotation of the actuator domain relative to the phosphorylation domain. Finally, bound phosphate at the reaction centre of the phosphorylation domain is hydrolysed owing to the displacement of the TGES loop. Because of the missing interaction between Ile199 and Met334 in their alanine-substituted mutants (Fig. 3c), the TM1–TM2 bundle may slip; therefore, the luminal gate closes spontaneously regardless of K+-binding to the cation-binding site. As a consequence, spontaneous dephosphorylation is induced, producing the K+-independent ATPase activity.
Extended Data Fig. 4 P-CAB-binding site.
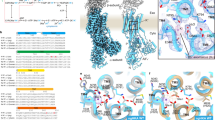
Inverse plot of 1/v versus 1/[K+] for the wild-type enzyme in the presence of different concentrations of P-CABs (vonoprazan: 0, 5, 10 and 20 nM (a); SCH28080: 0, 200, 500 and 1,000 nM (b), blue, green, yellow and red circles correspond to the respective P-CAB concentrations), showing typical K+-competitive inhibition of H+, K+-ATPase activity. Data represent mean ± s.e.m. of triplicated points at each of the indicated K+ concentrations; representative results from more than three independent measurements are shown. Their chemical structures are provided in each inset. c, d, The 2F o − F c electron density maps (contoured at 2σ) of the vonoprazan- (c) and SCH28080-binding site (d), viewed from approximately parallel to the membrane plane. In d, bound SCH28080 is depicted as wheat colour for clarity. e, f, Cross sections of the P-CAB-binding sites perpendicular to the membrane plane. The sectional surface is shown in light blue, and molecular surface is shown as light grey (carbon), with other colours corresponding to different elements (red, oxygen; blue, nitrogen; yellow, sulfur). Transparent spheres for each of the P-CABs represent their van der Waals radius, showing tight binding in their binding pocket. g. Structural comparison of the transmembrane region of vonoprazan-bound (magenta), SCH28080-bound (green) H+, K+-ATPase and ouabain-bound Na+, K+-ATPase (wheat), viewed from luminal side. Bound ouabain and Mg2+ ion in the Na+, K+-ATPase structure are shown for clarity. h, Ouabain and Mg2+ ion are superimposed on the vonoprazan-bound H+, K+-ATPase structure (ribbons). Seven amino acids of the H+, K+-ATPase, for which mutation provides high-affinity ouabain binding, are indicated (grey sticks), and their corresponding amino acids for Na+, K+-ATPase are indicated in parentheses. i, Bound SCH28080 is superimposed on the structure shown in h. See Supplementary Information for details.
Extended Data Fig. 5 F o − F c maps for P-CABs.
The F o − F c density for vonoprazan (a) and SCH28080 (b) contoured at 5σ (blue mesh) is shown in stereo view. The amino acids involved in the binding are indicated as sticks.
Extended Data Fig. 6 Cation-binding site in the Rb+-bound, luminal-open E2P state.
a, b, Close-up of the cation-binding site in H+, K+-ATPase (SCH)E2BeF viewed approximately perpendicular to the membrane from the cytoplasmic side (a) and parallel to the membrane from the TM4 side (b). Residues located within 3.5 Å between neighbouring atoms are connected by dotted lines. Bound Rb+ (purple sphere) and water molecules (red) are also indicated. c, d, Comparison of the cation-binding site between and Rb+-bound (SCH)E2BeF (colour ribbons) and (Von)E2BeF (wheat), showing the inclination of Glu820 side chain towards Rb+ accompanied by Rb+ binding (arrow). Only polar residues in the observed area are shown for clarity. e, f, K+-occluded (K+)2E2-MgF state of Na+, K+-ATPase (light grey, PDB code: 2ZXE) is superimposed on the Rb+-bound (SCH)E2BeF state of H+, K+-ATPase (colour ribbons). Pink spheres highlighted with red circles (site I and II) indicate bound K+ in the Na+, K+-ATPase structure. Atomic models are aligned based on the TM7–TM10 part of the proteins. Arrows indicate displacement of the TM4 luminal portion from the luminal-open to the luminal-closed form. TM5 is removed from the structures shown in d and f for clarity.
Extended Data Fig. 7 Hydrogen bond networks.
A transmembrane cation-binding site of H+, K+-ATPase (Von)E2BeF is shown, viewed from the TM6 side. Only polar residues are shown, and the distances between each residue are provided. Spheres indicate positions responsible for the Na+-binding site (I–III) in the Na+, K+-ATPase E1P-ADP state43. The proximity of Asp942 and Arg946 to one another indicates that these residues form a salt bridge.
Supplementary information
Supplementary Information
This file contains Supplementary Text and Supplementary References
Video 1: Crystal structure of H+,K+-ATPase bound to vonoprazan (Von)E2BeF
The atomic model of H+,K+-ATPase (Von)E2BeF as shown in Fig. 1a
Video 2: Cation-binding site in (Von)E2BeF
A transmembrane cation-binding site of H+,K+-ATPase (Von)E2BeF viewed parallel to the membrane normal with the cytoplasmic-side up. Polar side-chains likely to be involved in the cation transport are indicated with the distances between them
Video 3: Cation-binding site in Rb+-bound (SCH)E2BeF
A transmembrane cation-binding site of H+,K+-ATPase (SCH)E2BeF viewed parallel to the membrane normal with the cytoplasmic-side up. Bound Rb+ (purple sphere) and polar side-chains likely to be involved in the cation transport are indicated with the distances between them
Rights and permissions
About this article
Cite this article
Abe, K., Irie, K., Nakanishi, H. et al. Crystal structures of the gastric proton pump. Nature 556, 214–218 (2018). https://doi.org/10.1038/s41586-018-0003-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0003-8
This article is cited by
-
Amelioration of obsessive-compulsive disorder by intracellular acidification of cortical neurons with a proton pump inhibitor
Translational Psychiatry (2024)
-
Bioinformatic Study of Possible Acute Regulation of Acid Secretion in the Stomach
The Journal of Membrane Biology (2024)
-
Deep learning driven de novo drug design based on gastric proton pump structures
Communications Biology (2023)
-
Parkinson’s disease-associated ATP13A2/PARK9 functions as a lysosomal H+,K+-ATPase
Nature Communications (2023)
-
Electrostatic switch mechanisms of membrane protein trafficking and regulation
Biophysical Reviews (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.