Abstract
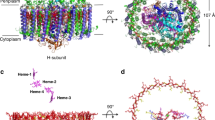
Light-harvesting complex 1 (LH1) and the reaction centre (RC) form a membrane-protein supercomplex that performs the primary reactions of photosynthesis in purple photosynthetic bacteria. The structure of the LH1–RC complex can provide information on the arrangement of protein subunits and cofactors; however, so far it has been resolved only at a relatively low resolution. Here we report the crystal structure of the calcium-ion-bound LH1–RC supercomplex of Thermochromatium tepidum at a resolution of 1.9 Å. This atomic-resolution structure revealed several new features about the organization of protein subunits and cofactors. We describe the loop regions of RC in their intact states, the interaction of these loop regions with the LH1 subunits, the exchange route for the bound quinone QB with free quinone molecules, the transport of free quinones between the inside and outside of the LH1 ring structure, and the detailed calcium-ion-binding environment. This structure provides a solid basis for the detailed examination of the light reactions that occur during bacterial photosynthesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cogdell, R. J. & Roszak, A. W. Structural biology: the purple heart of photosynthesis. Nature 508, 196–197 (2014).
Niwa, S. et al. Structure of the LH1–RC complex from Thermochromatium tepidum at 3.0Å. Nature 508, 228–232 (2014).
Yu, L. J., Kawakami, T., Kimura, Y. & Wang-Otomo, Z. Y. Structural basis for the unusual Qy red-shift and enhanced thermostability of the LH1 complex from Thermochromatium tepidum. Biochemistry 55, 6495–6504 (2016).
Qian, P. et al. Three-dimensional structure of the Rhodobacter sphaeroides RC-LH1-PufX complex: dimerization and quinone channels promoted by PufX. Biochemistry 52, 7575–7585 (2013).
Roszak, A. W. et al. Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science 302, 1969–1972 (2003).
Kimura, Y. et al. Calcium ions are involved in the unusual red shift of the light-harvesting 1 Qy transition of the core complex in thermophilic purple sulfur bacterium Thermochromatium tepidum. J. Biol. Chem. 283, 13867–13873 (2008).
Kimura, Y., Yu, L. J., Hirano, Y., Suzuki, H. & Wang, Z. Y. Calcium ions are required for the enhanced thermal stability of the light-harvesting-reaction center core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum. J. Biol. Chem. 284, 93–99 (2009).
Nogi, T., Fathir, I., Kobayashi, M., Nozawa, T. & Miki, K. Crystal structures of photosynthetic reaction center and high-potential iron-sulfur protein from Thermochromatium tepidum: thermostability and electron transfer. Proc. Natl Acad. Sci. USA 97, 13561–13566 (2000).
Roszak, A. W. et al. New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory. Biochem. J. 442, 27–37 (2012).
Weyer, K. A., Schafer, W., Lottspeich, F. & Michel, H. Cytochrome subunit of the photosynthetic reaction center from Rhodopseudomonas viridis is a lipoprotein. Biochemistry 26, 2909–2914 (1987).
Wöhri, A. B. et al. Lipidic sponge phase crystal structure of a photosynthetic reaction center reveals lipids on the protein surface. Biochemistry 48, 9831–9838 (2009).
Kulathila, R., Kulathila, R., Indic, M. & van den Berg, B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PLoS ONE 6, e15610 (2011).
Li, L. et al. Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 103, 19243–19248 (2006).
Kimura, Y. et al. Characterization of the quinones in purple sulfur bacterium Thermochromatium tepidum. FEBS Lett. 589, 1761–1765 (2015).
Wang-Otomo, Z.-Y. in Solar to Chemical Energy Conversion (eds Sugiyama, M. et al.) 379–390 (Springer International Publishing, Cham, 2016).
Stowell, M. H. B. et al. Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer. Science 276, 812–816 (1997).
Fritzsch, G., Kampmann, L., Kapaun, G. & Michel, H. Water clusters in the reaction centre of Rhodobacter sphaeroides. Photosynth. Res. 55, 127–132 (1998).
Abresch, E. C. et al. Identification of proton transfer pathways in the X-ray crystal structure of the bacterial reaction center from Rhodobacter sphaeroides. Photosynth. Res. 55, 119–125 (1998).
Krammer, E. M., Till, M. S., Sebban, P. & Ullmann, G. M. Proton-transfer pathways in photosynthetic reaction centers analyzed by profile hidden Markov models and network calculations. J. Mol. Biol. 388, 631–643 (2009).
Rücker, O., Köhler, A., Behammer, B., Sichau, K. & Overmann, J. Puf operon sequences and inferred structures of light-harvesting complexes of three closely related Chromatiaceae exhibiting different absorption characteristics. Arch. Microbiol. 194, 123–134 (2012).
Kimura, Y., Inada, Y., Yu, L. J., Wang, Z. Y. & Ohno, T. A spectroscopic variant of the light-harvesting 1 core complex from the thermophilic purple sulfur bacterium Thermochromatium tepidum. Biochemistry 50, 3638–3648 (2011).
Kimura, Y. et al. Metal cations modulate the bacteriochlorophyll–protein interaction in the light-harvesting 1 core complex from Thermochromatium tepidum. Biochim. Biophys. Acta 1817, 1022–1029 (2012).
Jakob-Grun, S., Radeck, J. & Braun, P. Ca2+-binding reduces conformational flexibility of RC-LH1 core complex from thermophile Thermochromatium tepidum. Photosynth. Res. 111, 139–147 (2012).
Ma, F., Yu, L. J., Wang-Otomo, Z. Y. & van Grondelle, R. The origin of the unusual Qy red shift in LH1–RC complexes from purple bacteria Thermochromatium tepidum as revealed by Stark absorption spectroscopy. Biochim. Biophys. Acta 1847, 1479–1486 (2015).
Ma, F., Yu, L. J., Wang-Otomo, Z. Y. & van Grondelle, R. Temperature dependent LH1 → RC energy transfer in purple bacteria Tch. tepidum with shiftable LH1-Qy band: a natural system to investigate thermally activated energy transfer in photosynthesis. Biochim. Biophys. Acta 1857, 408–414 (2016).
Ma, F. et al. Metal cations induced αβ-BChl a heterogeneity in LH1 as revealed by temperature-dependent fluorescence splitting. ChemPhysChem 18, 2295–2301 (2017).
Nagashima, K. V. P. et al. Probing structure–function relationships in early events in photosynthesis using a chimeric photocomplex. Proc. Natl Acad. Sci. USA 114, 10906–10911 (2017).
Suzuki, H. et al. Purification, characterization and crystallization of the core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum. Biochim. Biophys. Acta 1767, 1057–1063 (2007).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
The PyMOL Molecular Graphics System v.1.8 (Schrödinger, 2015).
Acknowledgements
We thank M. T. Madigan for providing the Tch. tepidum strain MC; F. Ma, Y. Xin, Y. Umena, X. Chen, X. Qin, W. Wang and T. Kawakami for discussion and assistance during the experiments and data analysis. This work was supported by JSPS KAKENHI No. JP24000018 and JP17H0643419 (to J.-R.S.), JP16H04174 (to Z.-Y.W.-O.), JP16H06296 and JP16H06162 (to M.S.), a program for promoting the enhancement of research universities at Okayama University from MEXT, Japan, and performed using the beamlines BL41XU (proposal numbers 2014B1277, 2015A1079, 2015B2079, 2016A2553, 2017A2590 to L.-J.Y.) and BL44XU (2015B6522, 2016A6621, 2016B6621, 2017A6724, 2017B6724 to M.S.) at SPring-8, and BL-1A at Photon Factory, Japan (2016R-27 to L.-J.Y.). We thank staff members of SPring-8 and Photon Factory for their assistance with data collection.
Reviewer information
Nature thanks R. Cogdell and R. Niederman for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
J.-R.S. and L.-J.Y. conceived the project; L.-J.Y. prepared the samples with the help of Z.-Y.W.-O.; L.-J.Y. grew the crystals under the supervision of J.-R.S.; L.-J.Y. and M.S. collected the diffraction data and analysed the structure; L.-J.Y. and J.-R.S. wrote the manuscript; and all authors contributed to the discussion and improvement of the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
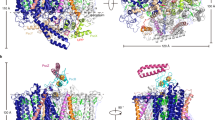
Extended Data Fig. 1 Quality of the LH1–RC crystal and its packing pattern.
a, An image of the LH1–RC crystals obtained in the present study. These crystals were obtained reproducibly under the present crystallization conditions. b, A typical diffraction image of the LH1–RC crystal taken at BL41XU of SPring-8, Japan, with a wavelength of 1.0 Å at 100 K. This diffraction image was obtained reproducibly with many crystals tested. c, d, Packing patterns of the previous (c) and the present crystal (d).
Extended Data Fig. 2 Close-up views of the electron density maps for some of the cofactors of LH1–RC.
The blue mesh represents the 2Fo−Fc map contoured at 1.0σ, taken at a wavelength of 1.0 Å and analysed to 1.9 Å resolution. a–d, The special-pair BChls (a), one pair of the LH1 BChls (b), one of the CDL (c) and the QB molecule (d).
Extended Data Fig. 3 Comparison of the arrangement of the cofactors between the previous and present structures.
a, Arrangement of the cofactors in the previous 3.0 Å structure, with a view from the top of the membrane. b, Superposition of the cofactors between the previous 3.0 Å and present 1.9 Å structures. c, The same as b, viewing from the side of the membrane. In b and c, the cofactors revealed in the present 1.9 Å structures are coloured differently, whereas those in the previous 3.0 Å structures are depicted in grey.
Extended Data Fig. 4 Comparison of the protein structures between the previous and present structures.
a, Superposition of the RC subunits between the previous 2.2 Å and the present 1.9 Å structures, with a side view from the membrane plane. b, Superposition of the RC subunits between the previous 3.0 Å and the present 1.9 Å structures, with a side view from the membrane plane. c, d, Superposition of the LH1 subunits between the previous 3.0 Å and present 1.9 Å structures, with a side view (c) and top view (d) relative to the membrane plane, respectively. In all panels, the present 1.9 Å structure is coloured, whereas the previous structures are depicted in grey.
Extended Data Fig. 5 Hydrogen-bonding networks for the protonation of QB.
a, Two possible proton channels connecting QB to the cytoplasmic surface. The thick arrow (coloured in blue) indicates the main channel formed within the H-subunit, which is enlarged in b, and the thin arrow indicates the second channel. b, The main hydrogen-bonding network indicated by the thick arrow in a, formed by a number of water molecules and the residues (green) from the H-subunit (pale cyan). QA and QB are depicted in violet and red, respectively, and the non-haem iron is depicted in deep purple. The hydrogen bonds are depicted as dashed lines. Water molecules participating in the hydrogen-bonding networks are depicted in orange, and those not participating are depicted in grey.
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, LJ., Suga, M., Wang-Otomo, ZY. et al. Structure of photosynthetic LH1–RC supercomplex at 1.9 Å resolution. Nature 556, 209–213 (2018). https://doi.org/10.1038/s41586-018-0002-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0002-9
This article is cited by
-
Bioinspired polymeric supramolecular columns as efficient yet controllable artificial light-harvesting platform
Nature Communications (2024)
-
High-resolution structure and biochemical properties of the LH1–RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum
Communications Biology (2024)
-
The synergy between the PscC subunits for electron transfer to the P840 special pair in Chlorobaculum tepidum
Photosynthesis Research (2024)
-
Atomic force microscopic analysis of the light-harvesting complex 2 from purple photosynthetic bacterium Thermochromatium tepidum
Photosynthesis Research (2023)
-
Role of hydrogen-bond networks on the donor side of photosynthetic reaction centers from purple bacteria
Biophysical Reviews (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.