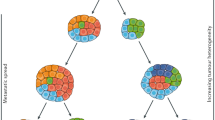
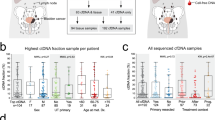
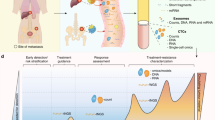
Abstract
Precision medicine has transformed the way urothelial carcinoma is managed. However, current practices are limited by the availability of tissue samples for genomic profiling and the spatial and temporal molecular heterogeneity observed in many studies. Among rapidly advancing genomic sequencing technologies, non-invasive liquid biopsy has emerged as a promising diagnostic tool to reproduce tumour genomics, and has shown potential to be integrated in several aspects of clinical care. In urothelial carcinoma, liquid biopsies such as plasma circulating tumour DNA (ctDNA) and urinary tumour DNA (utDNA) have been investigated as a surrogates for tumour biopsies and might bridge many shortfalls currently faced by clinicians. Both ctDNA and utDNA seem really promising in urothelial carcinoma diagnosis, staging and prognosis, response to therapy monitoring, detection of minimal residual disease and surveillance. The use of liquid biopsies in patients with urothelial carcinoma could further advance precision medicine in this population, facilitating personalized patient monitoring through non-invasive assays.
Key points
-
Genomic profiling in urothelial carcinoma has enabled precision medicine to transform the management of this malignancy.
-
Liquid biopsies are non-invasive genomic assays that serve as surrogates for the primary tumour biopsy. In urothelial carcinoma, liquid biopsies include urinary tumour DNA (utDNA) and circulating tumour DNA (ctDNA).
-
Advances in genomic sequencing techniques have enabled researchers to detect utDNA and ctDNA at previously undetectable levels. Genomic profiling of the primary tumour facilitates the creation of bespoke utDNA and ctDNA panels for patients with urothelial carcinoma.
-
In urothelial carcinoma, utDNA and ctDNA are very promising in multiple areas of care including diagnosis, risk stratification and prognostication, monitoring of response to systemic therapy, detection of minimal residual disease and surveillance.
-
Results from different studies have shown that utDNA and ctDNA outperform conventional markers for diagnosis and surveillance, showing promise for the integration of these factors into management paradigms. For example, utDNA and ctDNA consistently outperform urine cytology, and the detection of these markers identifies recurrence seen through cross-sectional imaging, holding implications for early and personalized systemic therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Change history
22 May 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41585-023-00783-6
References
Yoshida, T., Kates, M., Fujita, K., Bivalacqua, T. J. & McConkey, D. J. Predictive biomarkers for drug response in bladder cancer. Int. J. Urol. 26, 1044–1053 (2019).
Green, E. A. et al. Clinical utility of cell-free and circulating tumor DNA in kidney and bladder cancer: a critical review of current literature. Eur. Urol. Oncol. 4, 893–903 (2021).
Anker, P., Stroun, M. & Maurice, P. A. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 35, 2375–2382 (1975).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Ulz, P. et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 48, 1273–1278 (2016).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Utting, M., Werner, W., Dahse, R., Schubert, J. & Junker, K. Microsatellite analysis of free tumor DNA in urine, serum, and plasma of patients: a minimally invasive method for the detection of bladder cancer. Clin. Cancer Res. 8, 35–40 (2002).
Diaz, L. A. Jr & Bardelli, A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 32, 579–586 (2014).
Smith, A. B. et al. Muscle-invasive bladder cancer: evaluating treatment and survival in the National Cancer Data Base. BJU Int. 114, 719–726 (2014).
Kirkali, Z. et al. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology https://doi.org/10.1016/j.urology.2005.07.062 (2005).
Chamie, K. et al. Recurrence of high-risk bladder cancer: a population-based analysis. Cancer 119, 3219–3227 (2013).
Kompier, L. C. et al. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS ONE 5, e13821 (2010).
Pietzak, E. J. et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur. Urol. 72, 952–959 (2017).
Hosen, M. I. et al. Development of sensitive droplet digital PCR assays for detecting urinary TERT promoter mutations as non-invasive biomarkers for detection of urothelial cancer. Cancers 12, 3541 (2020).
Li, R. et al. Macroscopic somatic clonal expansion in morphologically normal human urothelium. Science 370, 82–89 (2020).
Lindskrog, S. V. et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat. Commun. 12, 2301 (2021).
Meeks, J. J. et al. Genomic characterization of high-risk non-muscle invasive bladder cancer. Oncotarget 7, 75176–75184 (2016).
Bacon, J. V. W. et al. Somatic features of response and relapse in non-muscle-invasive bladder cancer treated with bacillus Calmette-Guérin immunotherapy. Eur. Urol. Oncol. https://doi.org/10.1016/j.euo.2021.11.002 (2021).
Lamy, P. et al. Paired exome analysis reveals clonal evolution and potential therapeutic targets in urothelial carcinoma. Cancer Res. 76, 5894–5906 (2016).
Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014).
Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540–556.e25 (2017).
Trino, S. et al. P53-MDM2 pathway: evidences for a new targeted therapeutic approach in B-acute lymphoblastic leukemia. Front. Pharmacol. 7, 491 (2016).
Alhalabi, O. et al. Integrative clinical and genomic characterization of MTAP-deficient metastatic urothelial cancer. Eur. Urol. Oncol. https://doi.org/10.1016/j.euo.2021.10.006 (2021).
Glaser, A. P. et al. APOBEC-mediated mutagenesis in urothelial carcinoma is associated with improved survival, mutations in DNA damage response genes, and immune response. Oncotarget 9, 4537–4548 (2018).
Rosenberg, J. E. et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016).
Shohdy, K. S. et al. Serial ctDNA analysis predicts clinical progression in patients with advanced urothelial carcinoma. Br. J. Cancer 126, 430–439 (2022).
Ross, J. S. et al. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod. Pathol. 27, 271–280 (2014).
Witjes, J. A. et al. European Association of Urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur. Urol. 79, 82–104 (2021).
Plimack, E. R. et al. Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. Eur. Urol. 68, 959–967 (2015).
Van Allen, E. M. et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 4, 1140–1153 (2014).
Kufe D et al. (eds) Holland-Frei Cancer Medicine (BC Decker, 2003).
Groenendijk, F. H. et al. ERBB2 mutations characterize a subgroup of muscle-invasive bladder cancers with excellent response to neoadjuvant chemotherapy. Eur. Urol. 69, 384–388 (2016).
Park, C. M., Kawasaki, Y., Refaat, A. & Sakurai, H. Mechanisms for DNA-damaging agent-induced inactivation of ErbB2 and ErbB3 via the ERK and p38 signaling pathways. Oncol. Lett. 15, 1758–1762 (2018).
Gutierrez, C. & Schiff, R. HER2: biology, detection, and clinical implications. Arch. Pathol. Lab. Med. 135, 55–62 (2011).
Liu, D. et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat. Commun. 8, 2193 (2017).
Agarwal, N. et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer 124, 2115–2124 (2018).
Audenet, F. et al. Clonal relatedness and mutational differences between upper tract and bladder urothelial carcinoma. Clin. Cancer Res. 25, 967–976 (2019).
Fujii, Y. et al. Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer Cell 39, 793–809.e8 (2021).
Lee, J. K. et al. The pan-tumor landscape of targetable kinase fusions in circulating tumor DNA. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-21-2136 (2021).
Moss, T. J. et al. Comprehensive genomic characterization of upper tract urothelial carcinoma. Eur. Urol. 72, 641–649 (2017).
Nassar, A. H. et al. Mutational analysis of 472 urothelial carcinoma across grades and anatomic sites. Clin. Cancer Res. 25, 2458–2470 (2019).
Necchi, A. et al. A feasibility study of preoperative pembrolizumab before radical nephroureterectomy in patients with high-risk, upper tract urothelial carcinoma: PURE-02. Urol. Oncol. 40, 10.e1–10.e6 (2022).
Sfakianos, J. P. et al. Genomic characterization of upper tract urothelial carcinoma. Eur. Urol. 68, 970–977 (2015).
Su, X. et al. Comprehensive integrative profiling of upper tract urothelial carcinomas. Genome Biol. 22, 7 (2021).
Yang, K. et al. Comparison of genomic characterization in upper tract urothelial carcinoma and urothelial carcinoma of the bladder. Oncologist 26, e1395–e1405 (2021).
Donahu, T. F. et al. Genomic characterization of upper-tract urothelial carcinoma in patients with Lynch syndrome. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00143 (2018).
Bagrodia, A. et al. Genomic profile of urothelial carcinoma of the upper tract from ureteroscopic biopsy: feasibility and validation using matched radical nephroureterectomy specimens. Eur. Urol. Focus. 5, 365–368 (2019).
Wan, J. C. M. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Rodrigues Filho, E. M. et al. Elevated cell-free plasma DNA level as an independent predictor of mortality in patients with severe traumatic brain injury. J. Neurotrauma 31, 1639–1646 (2014).
Tsai, N.-W. et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin. Chim. Acta 412, 476–479 (2011).
Breitbach, S., Sterzing, B., Magallanes, C., Tug, S. & Simon, P. Direct measurement of cell-free DNA from serially collected capillary plasma during incremental exercise. J. Appl. Physiol. 117, 119–130 (2014).
De Vlaminck, I. et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl Acad. Sci. USA 112, 13336–13341 (2015).
Leon, S., Shapiro, B., Sklaroff, D. & Yaros, M. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 37, 646–650 (1977).
Sun, K. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl Acad. Sci. USA 112, E5503–E5512 (2015).
Diehl, F. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl Acad. Sci. USA 102, 16368–16373 (2005).
Dressman, D., Yan, H., Traverso, G., Kinzler, K. W. & Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA 100, 8817–8822 (2003).
Forshew, T. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 4, 136ra68 (2012).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Lebofsky, R. et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol. Oncol. 9, 783–790 (2015).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Heitzer, E. et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 5, 30 (2013).
Kirkizlar, E. et al. Detection of clonal and subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl. Oncol. 8, 407–416 (2015).
Sims, D., Sudbery, I., Ilott, N. E., Heger, A. & Ponting, C. P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 15, 121–132 (2014).
Botezatu, I. et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin. Chem. 461, 1078–1084 (2000).
Salvi, S. et al. The potential use of urine cell free DNA as a marker for cancer. Expert. Rev. Mol. Diagn. 16, 1283–1290 (2016).
Corcoran, R. B. & Chabner, B. A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 379, 1754–1765 (2018).
Birkenkamp-Demtröder, K. et al. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur. Urol. 70, 75–82 (2016).
Zhang, R. et al. Urinary molecular pathology for patients with newly diagnosed urothelial bladder cancer. J. Urol. 206, 873–884 (2021).
Satyal, U., Srivastava, A. & Abbosh, P. H. Urine biopsy-liquid gold for molecular detection and surveillance of bladder cancer. Front. Oncol. 9, 1266 (2019).
Hirotsu, Y. et al. Genomic profile of urine has high diagnostic sensitivity compared to cytology in non‐invasive urothelial bladder cancer. Cancer Sci. 110, 3235–3243 (2019).
Togneri, F. S. et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur. J. Hum. Genet. 24, 1167–1174 (2016).
Ou, Z. et al. Detection of bladder cancer using urinary cell-free DNA and cellular DNA. Clin. Transl. Med. 9, 4 (2020).
Jain, M. et al. Urine TERT promoter mutations-based tumor DNA detection in patients with bladder cancer: a pilot study. Mol. Clin. Oncol. 15, 253 (2021).
Avogbe, P. H. et al. Urinary TERT promoter mutations as non-invasive biomarkers for the comprehensive detection of urothelial cancer. EBioMedicine 44, 431–438 (2019).
Christensen, E. et al. Liquid biopsy analysis of FGFR3 and PIK3CA hotspot mutations for disease surveillance in bladder cancer. Eur. Urol. 71, 961–969 (2017).
Zhao, C. et al. A novel cell-free single-molecule unique primer extension resequencing (cf-SUPER) technology for bladder cancer non-invasive detection in urine. Transl. Androl. Urol. 9, 1222 (2020).
Hayashi, Y. et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell‐free DNA in upper tract urothelial carcinoma. Cancer Sci. 110, 1771–1779 (2019).
Grossfeld, G. D. et al. Evaluation of asymptomatic microscopic hematuria in adults: the American Urological Association best practice policy – part II: patient evaluation, cytology, voided markers, imaging, cystoscopy, nephrology evaluation, and follow-up. Urology 57, 604–610 (2001).
Dudley, J. C. et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov. 9, 500–509 (2019).
Witjes, J. A. et al. Performance of the bladder EpiCheck methylation test for patients under surveillance for non-muscle-invasive bladder cancer: results of a multicenter, prospective, blinded clinical trial. Eur. Urol. Oncol. 1, 307–313 (2018).
Pierconti, F. et al. Upper urothelial tract high-grade carcinoma: comparison of urine cytology and DNA methylation analysis in urinary samples. Hum. Pathol. 118, 42–48 (2021).
Chen, X. et al. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. J. Clin. Invest. 130, 6278–6289 (2020).
van Kessel, K. E. et al. Validation of a DNA methylation-mutation urine assay to select patients with hematuria for cystoscopy. J. Urol. 1971, 590–595 (2017).
Rai, B. P. et al. Systematic review of the incidence of and risk factors for urothelial cancers and renal cell carcinoma among patients with haematuria. Eur. Urol. https://doi.org/10.1016/j.eururo.2022.03.027 (2022).
Xu, Y. et al. A urine-based liquid biopsy method for detection of upper tract urinary carcinoma. Front. Oncol. 10, 597486 (2021).
Britton, J. P., Dowell, A. C. & Whelan, P. Dipstick haematuria and bladder cancer in men over 60: results of a community study. BMJ 299, 1010–1012 (1989).
Messing, E. M. et al. Comparison of bladder cancer outcome in men undergoing hematuria home screening versus those with standard clinical presentations. Urology 45, 387–396 (1995).
Barocas, D. A. et al. Microhematuria: AUA/SUFU guideline. J. Urol. 204, 778–786 (2020).
Xu, Y. et al. Diagnostic value of combined IQGAP3/BMP4 and IQGAP3/FAM107A expression ratios in urinary cell-free DNA for discriminating bladder cancer from hematuria. Urol. Oncol. 37, 86–96 (2019).
Cheng, T. H. T. et al. Noninvasive detection of bladder cancer by shallow-depth genome-wide bisulfite sequencing of urinary cell-free DNA for methylation and copy number profiling. Clin. Chem. 65, 927–936 (2019).
Humphrey, P. A., Moch, H., Cubilla, A. L., Ulbright, T. M. & Reuter, V. E. The 2016 WHO classification of tumours of the urinary system and male genital organs – part B: prostate and bladder tumours. Eur. Urol. 70, 106–119 (2016).
Kulkarni, G. S. et al. An updated critical analysis of the treatment strategy for newly diagnosed high-grade T1 (previously T1G3) bladder cancer. Eur. Urol. 57, 60–70 (2010).
Brausi, M. et al. Variability in the recurrence rate at first follow-up cystoscopy after TUR in stage Ta T1 transitional cell carcinoma of the bladder: a combined analysis of seven EORTC studies. Eur. Urol. 41, 523–531 (2002).
Kim, B. et al. Bladder tumor staging: comparison of contrast-enhanced CT, T1- and T2-weighted MR imaging, dynamic gadolinium-enhanced imaging, and late gadolinium-enhanced imaging. Radiology 193, 239–245 (1994).
Christensen, E. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
Zhang, Q. et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 10, 1842–1853 (2020).
Vandekerkhove, G. et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat. Commun. 12, 184 (2021).
Zhang, J. et al. Circulating tumor DNA analyses predict disease recurrence in non-muscle-invasive bladder cancer. Front. Oncol. 11, 657483 (2021).
Soave, A. et al. Copy number variations in primary tumor, serum and lymph node metastasis of bladder cancer patients treated with radical cystectomy. Sci. Rep. 10, 21562 (2020).
Grivas, P. et al. Circulating tumor DNA alterations in advanced urothelial carcinoma and association with clinical outcomes: a pilot study. Eur. Urol. Oncol. 3, 695–699 (2020).
Roupret, M. et al. European Association of Urology guidelines on upper urinary tract urothelial carcinoma: 2020 update. Eur. Urol. 79, 62–79 (2021).
Huelster, H. L. et al. Novel use of ctDNA to identify locally advanced and metastatic upper tract urothelial carcinoma [abstract]. J. Clin. Oncol. 40 (Suppl. 16), 4587 (2022).
de Almeida, E. F. et al. Plasma and urine DNA levels are related to microscopic hematuria in patients with bladder urothelial carcinoma. Clin. Biochem. 49, 1274–1277 (2016).
Patel, K. M. et al. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci. Rep. 7, 5554 (2017).
Pritchard, J. J. G. et al. Monitoring of urothelial cancer disease status after treatment by digital droplet PCR liquid biopsy assays. Urol. Oncol. 38, e10 (2020).
La Thangue, N. B. & Kerr, D. J. Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat. Rev. Clin. Oncol. 8, 587–596 (2011).
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403 (2017).
Reinert, T. et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131 (2019).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Chauhan, P. S. et al. Correction: Urine tumor DNA detection of minimal residual disease in muscle-invasive bladder cancer treated with curative-intent radical cystectomy: a cohort study. PLoS Med. 18, e1003876 (2021).
Gordon, N. S. et al. Urine DNA for monitoring chemoradiotherapy response in muscle-invasive bladder cancer: a pilot study. BJU Int. 129, 32–34 (2022).
Powles, T. et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 595, 432–437 (2021).
Powles, T. et al. An adaptive, biomarker-directed platform study of durvalumab in combination with targeted therapies in advanced urothelial cancer. Nat. Med. 27, 793–801 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04660344 (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04138628 (2022).
Vandekerkhove, G. et al. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin. Cancer Res. 23, 6487–6497 (2017).
Cescon, D. W., Bratman, S. V., Chan, S. M. & Siu, L. L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 1, 276–290 (2020).
Shen, J. et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 24, 556–562 (2018).
Kim, E. S. et al. Blood-based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: the phase 2 B-F1RST trial. Nat. Med. 28, 939–945 (2022).
Pal, S. K. et al. Efficacy of BGJ398, a fibroblast growth factor receptor 1–3 inhibitor, in patients with previously treated advanced urothelial carcinoma with FGFR3 alterations. Cancer Discov. 8, 812–821 (2018).
Ignatiadis, M., Sledge, G. W. & Jeffrey, S. S. Liquid biopsy enters the clinic – implementation issues and future challenges. Nat. Rev. Clin. Oncol. 18, 297–312 (2021).
Chan, H. T., Chin, Y. M., Nakamura, Y. & Low, S. K. Clonal hematopoiesis in liquid biopsy: from biological noise to valuable clinical implications. Cancers 12, 2277 (2020).
Crisafulli, G. et al. Temozolomide treatment alters mismatch repair and boosts mutational burden in tumor and blood of colorectal cancer patients. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-21-1434 (2022).
Barata, P. C. et al. Next-generation sequencing (NGS) of cell-free circulating tumor DNA and tumor tissue in patients with advanced urothelial cancer: a pilot assessment of concordance. Ann. Oncol. 28, 2458–2463 (2017).
Soave, A. et al. Copy number variations of circulating, cell-free DNA in urothelial carcinoma of the bladder patients treated with radical cystectomy: a prospective study. Oncotarget 8, 56398–56407 (2017).
Birkenkamp-Demtroder, K. et al. Monitoring treatment response and metastatic relapse in advanced bladder cancer by liquid biopsy analysis. Eur. Urol. 73, 535–540 (2018).
Raja, R. et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin. Cancer Res. 24, 6212–6222 (2018).
Borkowska, E. M. et al. Usefulness of droplet digital PCR and Sanger sequencing for detection of FGFR3 mutation in bladder cancer. Urol. Oncol. 37, 907–915 (2019).
Henriksen, T. V. et al. The effect of surgical trauma on circulating free DNA levels in cancer patients-implications for studies of circulating tumor DNA. Mol. Oncol. 14, 1670–1679 (2020).
Pal, S. K. et al. Infigratinib in upper tract urothelial carcinoma versus urothelial carcinoma of the bladder and its association with comprehensive genomic profiling and/or cell-free DNA results. Cancer 126, 2597–2606 (2020).
Blumendeller, C. et al. Use of plasma ctDNA as a potential biomarker for longitudinal monitoring of a patient with metastatic high-risk upper tract urothelial carcinoma receiving pembrolizumab and personalized neoepitope-derived multipeptide vaccinations: a case report. J. Immunother. Cancer https://doi.org/10.1136/jitc-2020-001406 (2021).
Chalfin, H. J. et al. Circulating tumor cell and circulating tumor DNA assays reveal complementary information for patients with metastatic urothelial cancer. Eur. Urol. Oncol. 4, 310–314 (2021).
Lee, D. H. et al. Urinary exosomal and cell-free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Sci. Rep. 8, 14707 (2018).
Casadio, V. et al. Urine cell-free DNA integrity as a marker for early bladder cancer diagnosis: preliminary data. Urol. Oncol. 31, 1744–1750 (2013).
Wang, K. et al. TERT promoter mutations are associated with distant metastases in upper tract urothelial carcinomas and serve as urinary biomarkers detected by a sensitive castPCR. Oncotarget 5, 12428–12439 (2014).
Kim, Y. H. et al. Value of urinary topoisomerase-IIA cell-free DNA for diagnosis of bladder cancer. Investig. Clin. Urol. 57, 106–112 (2016).
Casadio, V. et al. Cell-free DNA integrity analysis in urine samples. J. Vis. Exp. https://doi.org/10.3791/55049 (2017).
Kim, W. T. et al. Urinary cell-free nucleic acid IQGAP3: a new non-invasive diagnostic marker for bladder cancer. Oncotarget 9, 14354–14365 (2018).
Russo, I. J. et al. Toward personalised liquid biopsies for urothelial carcinoma: characterisation of ddPCR and urinary cfDNA for the detection of the TERT 228 G>A/T mutation. Bladder Cancer 4, 41–48 (2018).
Eich, M. L. et al. Incidence and distribution of UroSEEK gene panel in a multi-institutional cohort of bladder urothelial carcinoma. Mod. Pathol. 32, 1544–1550 (2019).
Hayashi, Y. et al. Clinical significance of hotspot mutation analysis of urinary cell-free DNA in urothelial bladder cancer. Front. Oncol. 10, 755 (2020).
Stasik, S. et al. Evaluation of TERT promoter mutations in urinary cell-free DNA and sediment DNA for detection of bladder cancer. Clin. Biochem. 64, 60–63 (2019).
Xu, Y. et al. Urinary cell-free DNA IQGAP3/BMP4 ratio as a prognostic marker for non-muscle-invasive bladder cancer. Clin. Genitourin. Cancer 17, e704–e711 (2019).
Ge, G. et al. Urothelial carcinoma detection based on copy number profiles of urinary cell-free DNA by shallow whole-genome sequencing. Clin. Chem. 66, 188–198 (2020).
Hentschel, A. E. et al. Comparative analysis of urine fractions for optimal bladder cancer detection using DNA methylation markers. Cancers 12, 859 (2020).
Lu, H. et al. Aristolochic acid mutational signature defines the low-risk subtype in upper tract urothelial carcinoma. Theranostics 10, 4323–4333 (2020).
Roperch, J. P. & Hennion, C. A novel ultra-sensitive method for the detection of FGFR3 mutations in urine of bladder cancer patients–design of the Urodiag(R) PCR kit for surveillance of patients with non-muscle-invasive bladder cancer (NMIBC). BMC Med. Genet. 21, 112 (2020).
Sieverink, C. A. et al. Clinical validation of a urine test (Uromonitor-V2((R))) for the surveillance of non-muscle-invasive bladder cancer patients. Diagnostics 10, 745 (2020).
Chauhan, P. S. et al. Urine tumor DNA detection of minimal residual disease in muscle-invasive bladder cancer treated with curative-intent radical cystectomy: a cohort study. PLoS Med. 18, e1003732 (2021).
Zhou, Z. et al. Jagged ends of urinary cell-free DNA: characterization and feasibility assessment in bladder cancer detection. Clin. Chem. 67, 621–630 (2021).
Author information
Authors and Affiliations
Contributions
K.M.R., H.L.H., P.E.S., R.K.J. and R.L. researched data for the article. K.M.R., H.L.H., J.J.M., B.M.F., S.P.L., P.E.S., R.K.J. and R.L. contributed substantially to discussion of the content. K.M.R., H.L.H., B.M.F., S.P.L., J.S.R., P.E.S., G.D.G., R.K.J., A.V. and R.L. wrote the article. K.M.R., H.L.H., J.J.M., B.M.F., G.P.S., S.P.L., J.S.R., G.D.G., A.M.K., A.V., L.W., X.W. and R.L. reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
K.M.R. is a consultant for Urogen Inc. J.J.M. is a consultant for Merck, AstraZeneca, Ferring, Incyte, Janssen, Foundation Medicine, BMS, UroGen, receives research funding from Epizyme, Hope Foundation, VHA, NIH, DoD; receives compensation for talks/educational courses from AUA, OncLive, Olympus, UroToday; clinical trials: SWOG, Genentech, Merck, AstraZeneca, Incyte; has two patents for T1 and TCGA classifier. B.M.F. is on the advisory board of Guardant, Janssen, Gilead, Merck, Immunomedics/Gilead, QED therapeutics; is a consultant for QED therapeutics, Boston gene; has patent royalties for Immunomedics/Gilead; receives honoraria from Urotoday; receives research support from Eli-Lilly. G.P.S. is an advisory board member for BMS, Genentech, EMD Serono, Merck, Sanofi, Seattle Genetics/Astellas, Astrazeneca, Exelixis, Janssen, Bicycle Therapeutics, Pfizer, Gilead, Scholar Rock, G1 Therapeutics, Eli Lilly/Loxo Oncology, Infinity Pharmaceuticals, Lucence Health, IMV; receives research support from Sanofi, Astrazeneca, Gilead, QED, Lucence, Predicine, BMS, EMD Serono, Jazz Therapeutics; is in the steering committee of the following studies: BMS, Bavarian Nordic, Seattle Genetics, QED, G1 Therapeutics (all unpaid), and Astrazeneca, EMD Serono, Debiopharm (paid); is on the data safety monitoring committee of Mereo; his spouse is employed by Myriad; travel costs: BMS, Astrazeneca; had writing/editor fees from Uptodate; is the Editor of Elsevier Practice Update Bladder Cancer Center of Excellence; receives speaking fees from Physicians Education Resource (PER), Onclive, Research to Practice, Medscape, Cancer Network, Masters Lecture Series (MLS). S.P.L. is a consultant/ advisory board member for Aura Bioscience, BMS, C2iGenomics, FerGene, Genentech, Merck, Pfizer/EMD Serono, Stimit, UroGen, Vaxiion, Verity; clinical trials: Endo, FKD, JBL (SWOG), Genentech (SWOG), QED Therapeutics, UroGen, Vaxiion, Viventia; has a patent for TCGA classifier; receives honoraria from Annenberg, Clinical Care Options, Grand Rounds Urology, Ology, UroToday. J.S.R. is an employee and equity holder in Foundation Medicine; is a consultant and equity holder in Tango Therapeutics and Celsius Therapeutics. P.E.S. is the NCCN bladder and penile cancer panel vice chair (leadership position, non-financially incentivized). R.K.J. is an advisory board member for Pfizer; is in the speakers bureau of Astellas/Seattle Genetics. A.M.K. is a consultant for Arquer Diagnostics, Asieris, Biological Dynamics, Bristol Myers Squibb, CG Oncology, H3 Biomedicine/Eisai, Engene, FerGene, Imagin Medical, Janssen, Medac, Merck, Photocure, ProTara, Seattle Genetics, Sessen Bio, Theralase, US Biotest, Urogen Inc, Roche, TMC Innovation; receives grants/research support from Adolor, BMS, FKD Industries, FerGene, Heat Biologics, Merck, Photocure, SWOG, NIH/GU SPORE, AIBCCR, Janssen (+ Taris), Seattle Genetics; has a patent for CyPRIT (Cytokine Predictors of Response to Intravesical Therapy); is a board member of the International Bladder Cancer Group (IBCG). R.L. receives research support from Predicine; Veracyte; CG Oncology; Valar labs; is in the clinical trial protocol committee of CG Oncology; is a scientific advisor/consultant for BMS, Merck, Fergene, Arquer Diagnostics, Urogen Pharma, Lucence, CG Oncology.
Peer review
Peer review information
Nature Reviews Urology thanks K. Junker and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rose, K.M., Huelster, H.L., Meeks, J.J. et al. Circulating and urinary tumour DNA in urothelial carcinoma — upper tract, lower tract and metastatic disease. Nat Rev Urol 20, 406–419 (2023). https://doi.org/10.1038/s41585-023-00725-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41585-023-00725-2