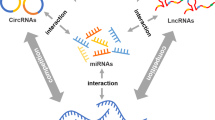
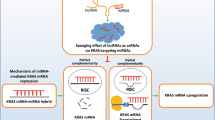
Abstract
Long non-coding RNAs (lncRNAs) are regulators of cellular machinery that are commonly dysregulated in genitourinary malignancies. Accordingly, the investigation of lncRNAs is improving our understanding of genitourinary cancers, from development to progression and dissemination. lncRNAs are involved in major oncogenic events in genitourinary malignancies, including androgen receptor (AR) signalling in prostate cancer, hypoxia-inducible factor (HIF) pathway activation in renal cell carcinoma and invasiveness in bladder cancer, as well as multiple other proliferation and survival mechanisms. In line with their putative oncogenic roles, new lncRNA-based classifications are emerging as potent predictors of prognosis. In clinical practice, detection of oncogenic lncRNAs in serum or urine might enable early cancer detection, and lncRNAs might also be promising therapeutic targets for patients with genitourinary cancer. Furthermore, as predictors of sensitivity to anticancer treatments, lncRNAs could be integrated into future precision medicine strategies. Overall, lncRNAs are promising new candidates for molecular studies and for discovery of innovative biomarkers and are putative therapeutic targets in genitourinary oncology.
Key points
-
Long non-coding RNAs (lncRNAs) are untranslated transcripts >200 nucleotides in length with a 3D structure that enables complex interactions with DNA, mRNA and proteins.
-
lncRNAs influence cellular functions through genome-wide transcriptional regulation as well as direct interactions with proteins in diverse signalling pathways.
-
lncRNAs are frequently dysregulated in genitourinary malignancies, resulting in the promotion of multiple oncogenic mechanisms and the acquisition of therapeutic resistance.
-
The detection of tissue-specific lncRNAs in liquid biopsy samples provides opportunities to improve the early diagnosis of genitourinary malignancies.
-
lncRNA expression signatures can improve the subtyping and risk classification of genitourinary cancers and, therefore, have the potential to help refine therapeutic strategies.
-
Targeting lncRNAs using mechanisms based on RNA interference might provide new therapeutic opportunities in genitourinary malignancies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014).
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Rini, B. et al. A 16-gene assay to predict recurrence after surgery in localised renal cell carcinoma: development and validation studies. Lancet Oncol. 16, 676–685 (2015).
Knowles, M. A. & Hurst, C. D. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 15, 25–41 (2015).
Beuselinck, B. et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin. Cancer Res. 21, 1329–1339 (2015).
Spratt, D. E., Zumsteg, Z. S., Feng, F. Y. & Tomlins, S. A. Translational and clinical implications of the genetic landscape of prostate cancer. Nat. Rev. Clin. Oncol. 13, 597–610 (2016).
Elaidi, R. T. et al. A phase 2 BIOmarker driven trial with Nivolumab and Ipilimumab or VEGFR tKi in naïve metastatic Kidney cancer: the BIONIKK trial [923TiP]. Ann. Oncol. 28, mdx371.077 (2017).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015). This paper details a landmark effort to characterize the lncRNA transcriptome in humans, from normal and tumour tissues to cell lines, identifying nearly 8,000 lineage-associated or cancer-associated lncRNAs.
Ogawa, Y., Sun, B. K. & Lee, J. T. Intersection of the RNAi and X-inactivation pathways. Science 320, 1336–1341 (2008). This study reports the characterization and regulation of the mechanisms of X inactivation by the lncRNA XIST , highlighting the essential role of non-coding transcripts in human biology.
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2016).
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Ransohoff, J. D., Wei, Y. & Khavari, P. A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell. Biol. 19, 143–157 (2018).
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R. & Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 19, 535–548 (2018).
Mercer, T. R. & Mattick, J. S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 (2013).
Novikova, I. V., Hennelly, S. P. & Sanbonmatsu, K. Y. Tackling structures of long noncoding RNAs. Int. J. Mol. Sci. 14, 23672–23684 (2013).
Ulitsky, I. & Bartel, D. P. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46 (2013).
Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011).
Guttman, M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009). This study reports that expression of lncRNAs is conserved in mammals and regulates key cellular functions through transcriptional regulation.
Zheng, G. X. Y., Do, B. T., Webster, D. E., Khavari, P. A. & Chang, H. Y. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat. Struct. Mol. Biol. 21, 585–590 (2014).
Marvin, M. C. et al. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA 17, 1441–1450 (2011).
Wilusz, J. E., Freier, S. M. & Spector, D. L. 3΄ end processing of a long nuclear-retained non-coding RNA yields a tRNA-like cytoplasmic RNA. Cell 135, 919–932 (2008).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Qu, L. et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016).
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018).
Flippot, R. et al. Cancer subtypes classification using long non-coding RNA. Oncotarget 7, 54082–54093 (2016).
Guttman, M. & Rinn, J. L. Modular regulatory principles of large non–coding RNAs. Nature 482, 339–346 (2012).
Geisler, S. & Coller, J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell. Biol. 14, 699–712 (2013).
Yoon, J.-H. et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 4, 2939 (2013).
Ørom, U. A. et al. Long non-coding RNAs with enhancer-like function in human. Cell 143, 46–58 (2010).
Long, Y., Wang, X., Youmans, D. T. & Cech, T. R. How do lncRNAs regulate transcription? Sci. Adv. 3, eaao2110 (2017).
Xiao, H. et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget 6, 38005–38015 (2015).
Chiu, H.-S. et al. Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 23, 297–312 (2018).
Romero-Barrios, N., Legascue, M. F., Benhamed, M., Ariel, F. & Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 46, 2169–2184 (2018).
Latos, P. A. et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 (2012).
Nishikura, K. A-To-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell. Biol. 17, 83–96 (2016).
Feng, J. et al. The RNA component of human telomerase. Science 269, 1236–1241 (1995).
Khalil, A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11667–11672 (2009). This study finds that multiple lncRNAs can interact physically with transcriptional regulators, such as PRC2, demonstrating their importance for the function of chromatin remodelling complexes.
Portoso, M. et al. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 36, 981–994 (2017).
Brockdorff, N. Noncoding RNA and Polycomb recruitment. RNA 19, 429–442 (2013).
Couso, J.-P. & Patraquim, P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell. Biol. 18, 575–589 (2017).
Martens-Uzunova, E. S. et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 65, 1140–1151 (2014).
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer J. Int. Cancer 136, E359–E386 (2015).
Groskopf, J. et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 52, 1089–1095 (2006).
Morris, M. J. et al. Optimizing anticancer therapy in metastatic non-castrate prostate cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 36, 1521–1539 (2018).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
Taylor, B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010).
Armenia, J. et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 (2018).
Smolle, M. A., Bauernhofer, T., Pummer, K., Calin, G. A. & Pichler, M. Current insights into long non-coding RNAs (lncRNAs) in prostate cancer. Int. J. Mol. Sci. 18, 473 (2017).
Takayama, K.-I. et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 32, 1665–1680 (2013).
Bussemakers, M. J. et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 59, 5975–5979 (1999). In this study, overexpression of PCA3 (also known as DD3 ) was identified as a sensitive and specific marker of prostate cancer, demonstrating the potential of lncRNAs for improving early diagnosis.
Ferreira, L. B. et al. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer 12, 507 (2012).
Lemos, A. E. G. et al. PCA3 long noncoding RNA modulates the expression of key cancer-related genes in LNCaP prostate cancer cells. Tumour Biol. 37, 11339–11348 (2016).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Zhang, A. et al. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 13, 209–221 (2015).
Yang, L. et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602 (2013).
Parolia, A. et al. The long non-coding RNA PCGEM1 is regulated by androgen receptor activity in vivo. Mol. Cancer 14, 46 (2015).
Cui, Z. et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol. Oncol. 31, 1117–1123 (2013).
Zhang, Y. et al. Analysis of the androgen receptor–regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat. Genet. 50, 814–824 (2018).
Salameh, A. et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl Acad. Sci. USA 112, 8403–8408 (2015).
Hung, C.-L. et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl Acad. Sci. USA 111, 18697–18702 (2014).
Prensner, J. R. et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia 16, 900–908 (2014).
Chakravarty, D. et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383 (2014).
Ricke, W. A. et al. Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. FASEB J. 22, 1512–1520 (2008).
Xiong, W. et al. Oncogenic non-coding RNA NEAT1 promotes the prostate cancer cell growth through the SRC3/IGF1R/AKT pathway. Int. J. Biochem. Cell Biol. 94, 125–132 (2018).
West, J. A. et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802 (2014).
Fox, A. H. & Lamond, A. I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2, a000687 (2010).
Prensner, J. R. et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 74, 1651–1660 (2014).
Li, Y. et al. Long noncoding RNA SChLAP1 accelerates the proliferation and metastasis of prostate cancer via targeting miR-198 and promoting the MAPK1 pathway. Oncol. Res. 26, 131–143 (2018).
Malek, R. et al. TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res. 77, 3181–3193 (2017).
Bauderlique-Le Roy, H. et al. Enrichment of human stem-like prostate cells with s-SHIP promoter activity uncovers a role in stemness for the long noncoding RNA H19. Stem Cells Dev. 24, 1252–1262 (2015).
Zhao, B. et al. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-β1/ Smad signaling pathway. Cancer Biomark. 21, 613–620 (2018).
Orfanelli, U. et al. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene 34, 2094–2102 (2015).
Wang, X., Yang, B. & Ma, B. The UCA1/miR-204/Sirt1 axis modulates docetaxel sensitivity of prostate cancer cells. Cancer Chemother. Pharmacol. 78, 1025–1031 (2016).
Byles, V. et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene 31, 4619–4629 (2012).
Poliseno, L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 (2010). In this paper, PTENP1 is shown to regulate the expression of PTEN, showing that lncRNAs can be potentially involved in the regulation of major oncogenic pathways .
Du, Z. et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun. 7, 10982 (2016).
Wang, X. et al. Long intragenic non-coding RNA lincRNA-p21 suppresses development of human prostate cancer. Cell Prolif. 50, e12318 (2017).
Wang, X. et al. LincRNA-p21 suppresses development of human prostate cancer through inhibition of PKM2. Cell Prolif. 50, e12395 (2017).
Du, Y. et al. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget 8, 94358–94370 (2017).
Li, J. et al. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol. Rep. 36, 3241–3250 (2016).
Pickard, M. R., Mourtada-Maarabouni, M. & Williams, G. T. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta 1832, 1613–1623 (2013).
Yacqub-Usman, K., Pickard, M. R. & Williams, G. T. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate 75, 693–705 (2015).
Liu, J. et al. Reciprocal regulation of long noncoding RNAs THBS4-003 and THBS4 control migration and invasion in prostate cancer cell lines. Mol. Med. Rep. 14, 1451–1458 (2016).
Jia, J. et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget 7, 37868–37881 (2016).
Sebastian, A., Hum, N. R., Hudson, B. D. & Loots, G. G. Cancer-osteoblast interaction reduces sost expression in osteoblasts and up-regulates lncRNA MALAT1 in prostate cancer. Microarrays 4, 503–519 (2015).
Wang, D. et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 6, 41045–41055 (2015).
Xu, K. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012).
Zhang, Y. et al. An androgen reduced transcript of LncRNA GAS5 promoted prostate cancer proliferation. PLOS ONE 12, e0182305 (2017).
Wang, G. et al. Lrf suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat. Genet. 45, 739–746 (2013).
Zhu, M. et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 281, 3766–3775 (2014).
Prensner, J. R. et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget 5, 1434–1438 (2014).
Prensner, J. R. et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392–1398 (2013).
Chua, M. L. K. et al. A prostate cancer ‘nimbosus’: genomic instability and SChLAP1 dysregulation underpin aggression of intraductal and cribriform subpathologies. Eur. Urol. 72, 665–674 (2017).
Hsieh, J. J. et al. Renal cell carcinoma. Nat. Rev. Dis. Primer 3, 17009 (2017).
Semenza, G. L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29, 625–634 (2010).
Brugarolas, J. PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer J. 19, 324–332 (2013).
Kanu, N. et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34, 5699–5708 (2015).
Malouf, G. G. et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Mol. Oncol. 9, 32–43 (2014). In this study, unsupervised clustering of the ccRCC transcriptome identifies four subgroups with distinct mutational and clinicopathological profiles, suggesting that lncRNAs can help define genitourinary tumour subtypes.
Wu, W. et al. Hypoxia induces H19 expression through direct and indirect Hif-1α activity, promoting oncogenic effects in glioblastoma. Sci. Rep. 7, 45029 (2017).
Wang, L. et al. Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma 62, 412–418 (2015).
He, H., Wang, N., Yi, X., Tang, C. & Wang, D. Long non-coding RNA H19 regulates E2F1 expression by competitively sponging endogenous miR-29a-3p in clear cell renal cell carcinoma. Cell Biosci. 7, 65 (2017).
Raveh, E., Matouk, I. J., Gilon, M. & Hochberg, A. The H19 long non-coding RNA in cancer initiation, progression and metastasis — a proposed unifying theory. Mol. Cancer 14, 184 (2015).
Zhang, H., Yang, F., Chen, S.-J., Che, J. & Zheng, J. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 36, 2947–2955 (2015).
Hirata, H. et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 75, 1322–1331 (2015).
Wu, Y. et al. Suppressed expression of long non-coding RNA HOTAIR inhibits proliferation and tumourigenicity of renal carcinoma cells. Tumour Biol. 35, 11887–11894 (2014).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by non-coding RNAs. Cell 129, 1311–1323 (2007).
Hong, Q. et al. LncRNA HOTAIR regulates HIF-1α/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 8, e2772 (2017).
Katayama, H. et al. Long non-coding RNA HOTAIR promotes cell migration by upregulating insulin growth factor–binding protein 2 in renal cell carcinoma. Sci. Rep. 7, 12016 (2017).
Ding, J. et al. Estrogen receptor β promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene 37, 5037–5053 (2018).
Zhai, W. et al. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2α/C-MYC axis under hypoxia. Oncogene 35, 4866–4880 (2016).
Qu, L. et al. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat. Commun. 7, 12692 (2016).
Wang, Y. et al. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am. J. Cancer Res. 7, 312–322 (2017).
Xu, Z. et al. Long noncoding RNA-SRLR elicits intrinsic sorafenib resistance via evoking IL-6/STAT3 axis in renal cell carcinoma. Oncogene 36, 1965–1977 (2017).
Gong, X. et al. Novel lincRNA SLINKY is a prognostic biomarker in kidney cancer. Oncotarget 8, 18657–18669 (2017).
Flippot, R. et al. Expression of long non-coding RNA MFI2-AS1 is a strong predictor of recurrence in sporadic localized clear-cell renal cell carcinoma. Sci. Rep. 7, 8540 (2017).
Thrash-Bingham, C. A. & Tartof, K. D. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J. Natl Cancer Inst. 91, 143–151 (1999).
Bertozzi, D. et al. Characterization of novel antisense HIF-1α transcripts in human cancers. Cell Cycle 10, 3189–3197 (2011).
Baranello, L., Bertozzi, D., Fogli, M. V., Pommier, Y. & Capranico, G. DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1alpha gene locus. Nucleic Acids Res. 38, 159–171 (2010).
Mineo, M. et al. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 15, 2500–2509 (2016).
Chen, M. et al. Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Cancer Lett. 376, 155–164 (2016).
Cayre, A., Rossignol, F., Clottes, E. & Penault-Llorca, F. aHIF but not HIF-1alpha transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 5, R223–R230 (2003).
Jiang, Y.-Z. et al. Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res. 76, 2105–2114 (2016).
Qiao, H.-P., Gao, W.-S., Huo, J.-X. & Yang, Z.-S. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac. J. Cancer Prev. 14, 1077–1082 (2013).
Zhou, Y. et al. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742 (2007).
Kawakami, T. et al. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Hum. Mol. Genet. 15, 821–830 (2006).
Yao, J. et al. Decreased expression of a novel lncRNA CADM1-AS1 is associated with poor prognosis in patients with clear cell renal cell carcinomas. Int. J. Clin. Exp. Pathol. 7, 2758–2767 (2014).
Xue, S. et al. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 8, 3765–3774 (2015).
Sanli, O. et al. Bladder cancer. Nat. Rev. Dis. Primer 3, 17022 (2017).
Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 171, 540–556 (2017).
Peter, S. et al. Identification of differentially expressed long noncoding RNAs in bladder cancer. Clin. Cancer Res. 20, 5311–5321 (2014).
Wang, L. et al. Genome-wide screening and identification of long noncoding RNAs and their interaction with protein coding RNAs in bladder urothelial cell carcinoma. Cancer Lett. 349, 77–86 (2014).
Wang, M. et al. Common and differentially expressed long noncoding RNAs for the characterization of high and low grade bladder cancer. Gene 592, 78–85 (2016).
Wang, H. et al. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget 7, 86174–86185 (2016).
Millán-Rodríguez, F., Chéchile-Toniolo, G., Salvador-Bayarri, J., Palou, J. & Vicente-Rodríguez, J. Multivariate analysis of the prognostic factors of primary superficial bladder cancer. J. Urol. 163, 73–78 (2000).
Wang, X.-S. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 12, 4851–4858 (2006).
Wang, F., Li, X., Xie, X., Zhao, L. & Chen, W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 582, 1919–1927 (2008).
Luo, J. et al. LncRNA UCA1 promotes the invasion and EMT of bladder cancer cells by regulating the miR-143/HMGB1 pathway. Oncol. Lett. 14, 5556–5562 (2017).
Wu, W. et al. Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLOS ONE 8, e73920 (2013).
Xue, M. et al. Upregulation of long non-coding RNA urothelial carcinoma associated 1 by CCAAT/enhancer binding protein α contributes to bladder cancer cell growth and reduced apoptosis. Oncol. Rep. 31, 1993–2000 (2014).
Xue, M., Li, X., Li, Z. & Chen, W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumor Biol. 35, 6901–6912 (2014).
Elkin, M. et al. The expression of the imprinted H19 and IGF-2 genes in human bladder carcinoma. FEBS Lett. 374, 57–61 (1995).
Ohana, P. et al. The expression of the H19 gene and its function in human bladder carcinoma cell lines. FEBS Lett. 454, 81–84 (1999).
Ariel, I. et al. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol. Pathol. 53, 320 (2000).
Luo, M. et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333, 213–221 (2013).
Lv, M. et al. lncRNA H19 regulates epithelial–mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim. Biophys. Acta 1864, 1887–1899 (2017).
Chen, L.-H., Hsu, W.-L., Tseng, Y.-J., Liu, D.-W. & Weng, C.-F. Involvement of DNMT 3B promotes epithelial-mesenchymal transition and gene expression profile of invasive head and neck squamous cell carcinomas cell lines. BMC Cancer 16, 431 (2016).
Luo, M. et al. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 280, 1709–1716 (2013).
Li, S. et al. The YAP1 oncogene contributes to bladder cancer cell proliferation and migration by regulating the H19 long noncoding RNA. Urol. Oncol. 33, 427.e1–427.e10 (2015).
Han, Y., Liu, Y., Nie, L., Gui, Y. & Cai, Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology 81, 209.e1–209.e7 (2013).
Fan, Y. et al. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 20, 1531–1541 (2014).
Ying, L. et al. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 8, 2289 (2012).
Tan, J., Qiu, K., Li, M. & Liang, Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 589, 3175–3181 (2015).
Han, Y., Liu, Y., Gui, Y. & Cai, Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder: TUG 1 in bladder cancer. J. Surg. Oncol. 107, 555–559 (2013).
Zhuang, J. et al. TGFβ1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci. Rep. 5, 11924 (2015).
Li, L.-J. et al. Long noncoding RNA GHET1 promotes the development of bladder cancer. Int. J. Clin. Exp. Pathol. 7, 7196 (2014).
Berrondo, C. et al. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLOS ONE 11, e0147236 (2016).
Droop, J. et al. Diagnostic and prognostic value of long noncoding RNAs as biomarkers in urothelial carcinoma. PLOS ONE 12, e0176287 (2017).
Li, Z., Li, X., Wu, S., Xue, M. & Chen, W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 105, 951–955 (2014).
Li, H.-J. et al. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn J. Clin. Oncol. 45, 1055–1063 (2015).
Wang, X. et al. Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncol. Rep. 32, 1281–1290 (2014).
Yang, C., Li, X., Wang, Y., Zhao, L. & Chen, W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene 496, 8–16 (2012).
Liu, C. et al. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumor Biol. 37, 263–270 (2016).
Sun, X. et al. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 6, e1907 (2015).
Xiong, Y. et al. The long non-coding RNA XIST interacted with MiR-124 to modulate bladder cancer growth, invasion and migration by targeting androgen receptor (AR). Cell. Physiol. Biochem. 43, 405–418 (2017).
Liu, Z. et al. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLOS ONE 8, e73991 (2013).
Cao, Q., Wang, N., Qi, J., Gu, Z. & Shen, H. Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (C-C motif) ligand 1 expression. Mol. Med. Rep. 13, 27–34 (2016).
Ying, L. et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. Biosyst. 9, 407 (2013).
Qi, P., Zhou, X. & Du, X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol. Cancer 15, 39 (2016).
Jiang, H., Hu, X., Zhang, H. & Li, W. Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat. Oncol. 12, 65 (2017).
Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 12, 433–446 (2013).
Han, Y. et al. Prostate cancer susceptibility in men of African ancestry at 8q24. J. Natl. Cancer Inst. 108, djv431 (2016).
Verhaegh, G. W. et al. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur. Urol. 54, 1118–1126 (2008).
Hua, Q. et al. Genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population. Mutagenesis 31, 531–538 (2016).
Wang, F. et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget 5, 11091–11102 (2014).
Zhang, W. et al. A novel urinary long non-coding RNA transcript improves diagnostic accuracy in patients undergoing prostate biopsy. Prostate 75, 653–661 (2015).
de Kok, J. B. et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 62, 2695–2698 (2002).
Hessels, D. et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44, 8–15 (2003).
Haese, A. et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur. Urol. 54, 1081–1088 (2008).
Sanguedolce, F. et al. Urine TMPRSS2: ERG fusion transcript as a biomarker for prostate cancer: literature review. Clin. Genitourin. Cancer 14, 117–121 (2016).
Ke, D. et al. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget 8, 21516–21525 (2017).
Permuth, J. B. et al. Linc-ing circulating long non-coding RNAs to the diagnosis and malignant prediction of intraductal papillary mucinous neoplasms of the pancreas. Sci. Rep. 7, 10484 (2017).
Ren, S. et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 49, 2949–2959 (2013).
Li, M. et al. Long non-coding RNAs in renal cell carcinoma: a systematic review and clinical implications. Oncotarget 8, 48424–48435 (2017).
Wu, Y. et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis 5, e192 (2016).
Wang, Z. et al. Long non-coding RNA urothelial carcinoma–associated 1 as a tumor biomarker for the diagnosis of urinary bladder cancer. Tumor Biol. 39, 1010428317709990 (2017).
Srivastava, A. K. et al. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour Biol. 35, 11435–11442 (2014).
Eissa, S., Matboli, M., Essawy, N. O. E. & Kotb, Y. M. Integrative functional genetic-epigenetic approach for selecting genes as urine biomarkers for bladder cancer diagnosis. Tumor Biol. 36, 9545–9552 (2015).
Duan, W. et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 7, 78850 (2016).
Ylipää, A. et al. Transcriptome sequencing reveals PCAT5 as a novel ERG-regulated long noncoding RNA in prostate cancer. Cancer Res. 75, 4026–4031 (2015).
Ma, W. et al. The prognostic value of long noncoding RNAs in prostate cancer: a systematic review and meta-analysis. Oncotarget 8, 57755–57765 (2017).
Mehra, R. et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur. Urol. 70, 549–552 (2016).
Prensner, J. R. et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 15, 1469–1480 (2014).
Huang, T.-B. et al. A potential panel of four-long noncoding RNA signature in prostate cancer predicts biochemical recurrence-free survival and disease-free survival. Int. Urol. Nephrol. 49, 825–835 (2017).
Liu, D. et al. The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget 7, 57228–57238 (2016).
Cheng, W.-S. et al. Both genes and lncRNAs can be used as biomarkers of prostate cancer by using high throughput sequencing data. Eur. Rev. Med. Pharmacol. Sci. 18, 3504–3510 (2014).
Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Zigeuner, R. et al. External validation of the Mayo Clinic Stage, Size, Grade, and Necrosis (SSIGN) score for clear-cell renal cell carcinoma in a single European centre applying routine pathology. Eur. Urol. 57, 102–111 (2010).
Leibovich, B. C. et al. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer 97, 1663–1671 (2003).
Qu, L. et al. Prognostic value of a long non-coding RNA signature in localized clear cell renal cell carcinoma. Eur. Urol. 74, 756–763 (2018). In this paper, lncRNA-based expression signatures are shown to be independent biomarkers for patient stratification in ccRCC.
Zhang, H.-M., Yang, F.-Q., Yan, Y., Che, J.-P. & Zheng, J.-H. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 7, 5801–5809 (2014).
Xiong, J., Liu, Y., Jiang, L., Zeng, Y. & Tang, W. High expression of long non-coding RNA lncRNA-ATB is correlated with metastases and promotes cell migration and invasion in renal cell carcinoma. Jpn J. Clin. Oncol. 46, 378–384 (2016).
Song, S. et al. RCCRT1 is correlated with prognosis and promotes cell migration and invasion in renal cell carcinoma. Urology 84, 730.e1–730.e7 (2014).
Su, H. et al. Decreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinoma. Oncotarget 8, 5789–5799 (2017).
Jin, P., Wang, J. & Liu, Y. Downregulation of a novel long non-coding RNA, LOC389332, is associated with poor prognosis and tumor progression in clear cell renal cell carcinoma. Exp. Ther. Med. 13, 1137–1142 (2017).
Hedegaard, J. et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 30, 27–42 (2016).
Yan, T.-H. et al. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumor Biol. 35, 10249–10257 (2014).
Zhang, S. et al. lncRNA up-regulated in nonmuscle invasive bladder cancer facilitates tumor growth and acts as a negative prognostic factor of recurrence. J. Urol. 196, 1270–1278 (2016).
Chen, T. et al. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem. Biophys. Res. Commun. 468, 666–670 (2015).
Gu, P. et al. lncRNA HOXD-AS1 regulates proliferation and chemo-resistance of castration-resistant prostate cancer via recruiting WDR5. Mol. Ther. 25, 1959–1973 (2017). This study reports that lncRNAs can drive resistance to therapy in genitourinary cancers, as demonstrated by HOXD-AS1 , which promotes resistance to paclitaxel and anti-androgens in prostate cancer models.
Misawa, A., Takayama, K.-I., Urano, T. & Inoue, S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J. Biol. Chem. 291, 17861–17880 (2016).
Ren, S. et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 190, 2278–2287 (2013).
Xue, D., Lu, H., Xu, H.-Y., Zhou, C.-X. & He, X.-Z. Long noncoding RNA MALAT1 enhances the docetaxel resistance of prostate cancer cells via miR-145-5p-mediated regulation of AKAP12. J. Cell. Mol. Med. 22, 3223–3237 (2018).
Fotouhi Ghiam, A. et al. Long non-coding RNA urothelial carcinoma associated 1 (UCA1) mediates radiation response in prostate cancer. Oncotarget 8, 4668–4689 (2017).
Wang, Y., van Boxel-Dezaire, A. H. H., Cheon, H., Yang, J. & Stark, G. R. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc. Natl Acad. Sci. USA 110, 16975–16980 (2013).
Fan, Y. et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 281, 1750–1758 (2014).
Shang, C., Guo, Y., Zhang, H. & Xue, Y. Long noncoding RNA HOTAIR is a prognostic biomarker and inhibits chemosensitivity to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother. Pharmacol. 77, 507–513 (2016).
Zhang, H., Guo, Y., Song, Y. & Shang, C. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother. Pharmacol. 79, 49–55 (2017).
Katayama, S. et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 (2005).
Wahlestedt, C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov. Today 11, 503–508 (2006).
Modarresi, F. et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459 (2012). This paper shows that targeting antisense transcripts, including lncRNAs, with single-stranded oligonucleotides can alter gene expression and can be used as a therapeutic tool in cancer.
Liang, X.-H., Sun, H., Nichols, J. G. & Crooke, S. T. RNase H1-dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Mol. Ther. 25, 2075–2092 (2017).
Huang, L. & Liu, Y. In vivo delivery of RNAi with lipid-based nanoparticles. Annu. Rev. Biomed. Eng. 13, 507–530 (2011).
Shim, M. S. & Kwon, Y. J. Efficient and targeted delivery of siRNA in vivo. FEBS J. 277, 4814–4827 (2010).
Johnson, D. B., Puzanov, I. & Kelley, M. C. Talimogene laherparepvec (T-VEC) for the treatment of advanced melanoma. Immunotherapy 7, 611–619 (2015).
Bennett, C. F. & Swayze, E. E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293 (2010).
Senn, J. J., Burel, S. & Henry, S. P. Non-CpG-containing antisense 2΄-methoxyethyl oligonucleotides activate a proinflammatory response independent of Toll-like receptor 9 or myeloid differentiation factor 88. J. Pharmacol. Exp. Ther. 314, 972–979 (2005).
Fu, X. et al. Synthetic artificial microRNAs targeting UCA1-MALAT1 or c-Myc inhibit malignant phenotypes of bladder cancer cells T24 and 5637. Mol. Biosyst. 11, 1285–1289 (2015).
Sidi, A. A. et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J. Urol. 180, 2379–2383 (2008).
Gofrit, O. N. et al. DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J. Urol. 191, 1697–1702 (2014).
Halachmi, S. et al. Phase II trial of BC-819 intravesical gene therapy in combination with BCG in patients with non-muscle invasive bladder cancer (NMIBC). J. Clin. Oncol. 36, 499–499 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03719300 (2019).
Chen, F. et al. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 14, 2476–2489 (2016).
Park, J. et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018).
Reviewer information
Nature Reviews Urology thanks I. Seim, J. Schalken and other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to this manuscript.
Corresponding author
Ethics declarations
Competing interests
G.G.M. has received consultancy fees from Bristol-Myers Squibb, Pfizer, Novartis and Ipsen. R.F. has received travel grants from Novartis and Pfizer. The other authors declare no competing interests related to this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Flippot, R., Beinse, G., Boilève, A. et al. Long non-coding RNAs in genitourinary malignancies: a whole new world. Nat Rev Urol 16, 484–504 (2019). https://doi.org/10.1038/s41585-019-0195-1
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41585-019-0195-1
This article is cited by
-
The impact of long non-coding RNA H19 on metabolic features and reproductive phenotypes of Egyptian women with polycystic ovary syndrome
Middle East Fertility Society Journal (2024)
-
ANRIL, H19 and TUG1: a review about critical long non-coding RNAs in cardiovascular diseases
Molecular Biology Reports (2024)
-
NEAT1 promotes the progression of prostate cancer by targeting the miR-582-5p/EZH2 regulatory axis
Cytotechnology (2024)
-
GATA6-AS1 suppresses epithelial–mesenchymal transition of pancreatic cancer under hypoxia through regulating SNAI1 mRNA stability
Journal of Translational Medicine (2023)
-
ADAMTS9-AS1 inhibits tumor growth and drug resistance in clear cell renal cell carcinoma via recruiting HuR to enhance ADAMTS9 mRNA stability
Cancer Nanotechnology (2023)