Abstract
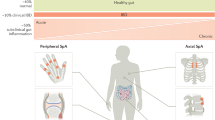
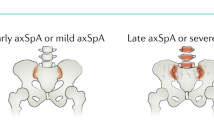
The term axial spondyloarthritis (axSpA) encompasses a heterogeneous group of diseases that have variable presentations, extra-articular manifestations and clinical outcomes, and that will respond differently to treatments. The prototypical type of axSpA, ankylosing spondylitis, is thought to be caused by interaction between the genetically primed host immune system and gut microbiota. Currently used biomarkers such as HLA-B27 status, C-reactive protein and erythrocyte sedimentation rate have, at best, moderate diagnostic and predictive value. Improved biomarkers are needed for axSpA to assist with early diagnosis and to better predict treatment responses and long-term outcomes. Advances in a range of ‘omics’ technologies and statistical approaches, including genomics approaches (such as polygenic risk scores), microbiome profiling and, potentially, transcriptomic, proteomic and metabolomic profiling, are making it possible for more informative biomarker sets to be developed for use in such clinical applications. Future developments in this field will probably involve combinations of biomarkers that require novel statistical approaches to analyse and to produce easy to interpret metrics for clinical application. Large publicly available datasets from well-characterized case–cohort studies that use extensive biological sampling, particularly focusing on early disease and responses to medications, are required to establish successful biomarker discovery and validation programmes.
Key points
-
Genomic and proteomic biomarkers in current clinical use for axial spondyloarthritis (axSpA) perform moderately well but there is a great need for more informative biomarkers.
-
Polygenic risk scores capture a greater proportion of the genetic component of risk of ankylosing spondylitis and perform better than HLA-B27 testing for the diagnosis of this disease.
-
Multiomic biomarkers are an underexplored area in axSpA that have the potential to be more informative than individual biomarkers.
-
Future biomarker development requires the availability of biological samples from large, well-characterized patient cohorts and datasets.
-
Biomarker development programmes should be an integral component of clinical trials, and biomarker data from those trials should be publicly available for biomarker development research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Poddubnyy, D. et al. Evaluation of 2 screening strategies for early identification of patients with axial spondyloarthritis in primary care. J. Rheumatol. 38, 2452–2460 (2011).
Bohn, R., Cooney, M., Deodhar, A., Curtis, J. R. & Golembesky, A. Incidence and prevalence of axial spondyloarthritis: methodologic challenges and gaps in the literature. Clin. Exp. Rheumatol. 36, 263–274 (2018).
Haroon, N. et al. The impact of tumor necrosis factor alpha inhibitors on radiographic progression in ankylosing spondylitis. Arthritis Rheum. 65, 2645–2654 (2013).
Maas, F. et al. Reduction in spinal radiographic progression in ankylosing spondylitis patients receiving prolonged treatment with tumor necrosis factor inhibitors. Arthritis Care Res. 69, 1011–1019 (2017).
Choy, E. et al. The need for comparative data in spondyloarthritis. Arthritis Res. Ther. 21, 32 (2019).
Feldtkeller, E., Khan, M. A., van der Heijde, D., van der Linden, S. & Braun, J. Age at disease onset and diagnosis delay in HLA-B27 negative vs. positive patients with ankylosing spondylitis. Rheumatol. Int. 23, 61–66 (2003).
Reed, M. D. et al. Ankylosing spondylitis: an Australian experience. Intern. Med. J. 38, 321–327 (2008).
Garrido-Cumbrera, M. et al. The European map of axial spondyloarthritis: capturing the patient perspective — an analysis of 2846 patients across 13 countries. Curr. Rheumatol. Rep. 21, 19 (2019).
Rudwaleit, M. et al. Effectiveness, safety, and predictors of good clinical response in 1250 patients treated with adalimumab for active ankylosing spondylitis. J. Rheumatol. 36, 801–808 (2009).
van der Linden, S., Valkenburg, H. & Cats, A. The risk of developing ankylosing spondylitis in HLA-B27 positive individuals: a family and population study. Br. J. Rheumatol. 22, 18–19 (1983).
Aletaha, D. & Smolen, J. S. Diagnosis and management of rheumatoid arthritis: a review. JAMA 320, 1360–1372 (2018).
Rudwaleit, M. et al. The development of Assessment of SpondyloArthritis International Society classification criteria for axial spondyloarthritis (part I): classification of paper patients by expert opinion including uncertainty appraisal. Ann. Rheum. Dis. 68, 770–776 (2009).
Jones, G. T., Dean, L. E., Pathan, E., Hollick, R. J. & Macfarlane, G. J. Real-world evidence of TNF inhibition in axial spondyloarthritis: can we generalise the results from clinical trials? Ann. Rheum. Dis. 79, 914–919 (2020).
Zhao, S. S. et al. Smoking status and cause-specific discontinuation of tumour necrosis factor inhibitors in axial spondyloarthritis. Arthritis Res. Ther. 21, 177 (2019).
Weber, U. & Baraliakos, X. Imaging in axial spondyloarthritis: changing concepts and thresholds. Best. Pract. Res. Clin. Rheumatol. 32, 342–356 (2018).
Kucybala, I., Urbanik, A. & Wojciechowski, W. Radiologic approach to axial spondyloarthritis: where are we now and where are we heading? Rheumatol. Int. 38, 1753–1762 (2018).
Rudwaleit, M., van der Heijde, D., Khan, M. A., Braun, J. & Sieper, J. How to diagnose axial spondyloarthritis early. Ann. Rheum. Dis. 63, 535–543 (2004).
Brown, M. A. et al. Evaluation of the effect of baseline MRI sacroiliitis and C reactive protein status on etanercept treatment response in non-radiographic axial spondyloarthritis: a post hoc analysis of the EMBARK study. Ann. Rheum. Dis. 77, 1091–1093 (2018).
Baraliakos, X., Szumski, A., Koenig, A. S. & Jones, H. The role of C-reactive protein as a predictor of treatment response in patients with ankylosing spondylitis. Semin. Arthritis Rheum. 48, 997–1004 (2019).
Van den Bosch, F. et al. Clinical and quality of life improvements with golimumab or infliximab in a real-life ankylosing spondylitis population: the QUO-VADIS study. Clin. Exp. Rheumatol. 37, 199–207 (2019).
Siebuhr, A. S., Bay-Jensen, A. C., Karsdal, M. A., Lories, R. J. & de Vlam, K. CRP and a biomarker of type I collagen degradation, C1M, can differentiate anti-inflammatory treatment response in ankylosing spondylitis. Biomark. Med. 10, 197–208 (2016).
Sieper, J. et al. Efficacy and safety of adalimumab in patients with non-radiographic axial spondyloarthritis: results of a randomised placebo-controlled trial (ABILITY-1). Ann. Rheum. Dis. 72, 815–822 (2013).
Braun, J., Baraliakos, X., Hermann, K. G., Xu, S. & Hsu, B. Serum C-reactive protein levels demonstrate predictive value for radiographic and magnetic resonance imaging outcomes in patients with active ankylosing spondylitis treated with golimumab. J. Rheumatol. 43, 1704–1712 (2016).
de Vries, M. K. et al. Erythrocyte sedimentation rate, C-reactive protein level, and serum amyloid a protein for patient selection and monitoring of anti-tumor necrosis factor treatment in ankylosing spondylitis. Arthritis Rheum. 61, 1484–1490 (2009).
Spoorenberg, A. et al. Relative value of erythrocyte sedimentation rate and C-reactive protein in assessment of disease activity in ankylosing spondylitis. J. Rheumatol. 26, 980–984 (1999).
Ruof, J. & Stucki, G. Validity aspects of erythrocyte sedimentation rate and C-reactive protein in ankylosing spondylitis: a literature review. J. Rheumatol. 26, 966–970 (1999).
Tsang, H. H. L. & Chung, H. Y. The discriminative values of the Bath Ankylosing Spondylitis Disease Activity Index, Ankylosing Spondylitis Disease Activity Score, C-reactive protein, and erythrocyte sedimentation rate in spondyloarthritis-related axial arthritis. J. Clin. Rheumatol. 23, 267–272 (2017).
Machado, P. et al. MRI inflammation and its relation with measures of clinical disease activity and different treatment responses in patients with ankylosing spondylitis treated with a tumour necrosis factor inhibitor. Ann. Rheum. Dis. 71, 2002–2005 (2012).
Bredella, M. A., Steinbach, L. S., Morgan, S., Ward, M. & Davis, J. C. MRI of the sacroiliac joints in patients with moderate to severe ankylosing spondylitis. AJR Am. J. Roentgenol. 187, 1420–1426 (2006).
Pedersen, S. J. et al. Radiographic progression is associated with resolution of systemic inflammation in patients with axial spondylarthritis treated with tumor necrosis factor alpha inhibitors: a study of radiographic progression, inflammation on magnetic resonance imaging, and circulating biomarkers of inflammation, angiogenesis, and cartilage and bone turnover. Arthritis Rheum. 63, 3789–3800 (2011).
Przepiera-Bedzak, H., Fischer, K. & Brzosko, M. Extra-articular symptoms in constellation with selected serum cytokines and disease activity in spondyloarthritis. Mediators Inflamm. 2016, 7617954 (2016).
Essers, I. et al. Characteristics associated with the presence and development of extra-articular manifestations in ankylosing spondylitis: 12-year results from OASIS. Rheumatology 54, 633–640 (2015).
Brown, M. A. et al. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum. 40, 1823–1828 (1997).
Pedersen, O. B. et al. Ankylosing spondylitis in Danish and Norwegian twins: occurrence and the relative importance of genetic vs. environmental effectors in disease causation. Scand. J. Rheumatol. 37, 120–126 (2008).
Ellinghaus, D. et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48, 510–518 (2016).
Cortes, A. et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat. Commun. 6, 7146 (2015).
Hill, A. V. et al. HLA class I typing by PCR: HLA-B27 and an African B27 subtype. Lancet 337, 640–642 (1991).
Braun, J. et al. Prevalence of spondylarthropathies in HLA-B27 positive and negative blood donors. Arthritis Rheum. 41, 58–67 (1998).
Jaakkola, E. et al. Finnish HLA studies confirm the increased risk conferred by HLA-B27 homozygosity in ankylosing spondylitis. Ann. Rheum. Dis. 65, 775–780 (2006).
Khan, M., Kushner, I., Braun, W., Zachary, A. & Steinberg, A. HLA-B27 homozygosity in ankylosing spondylitis: relationship to risk and severity. Tissue Antigens 11, 434–438 (1978).
International Genetics of Ankylosing Spondylitis Consortium. et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 (2013).
Hamersma, J. et al. Is disease severity in ankylosing spondylitis genetically determined? Arthritis Rheum. 44, 1396–1400 (2001).
Robinson, P. C. et al. Genetic dissection of acute anterior uveitis reveals similarities and differences in associations observed with ankylosing spondylitis. Arthritis Rheumatol. 67, 140–151 (2015).
Chung, H. Y., Machado, P., van der Heijde, D., D’Agostino, M. A. & Dougados, M. HLA-B27 positive patients differ from HLA-B27 negative patients in clinical presentation and imaging: results from the DESIR cohort of patients with recent onset axial spondyloarthritis. Ann. Rheum. Dis. 70, 1930–1936 (2011).
Okada, Y. et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am. J. Hum. Genet. 95, 162–172 (2014).
Goyette, P. et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet. 47, 172–179 (2015).
Cortes, A. et al. Association study of genes related to bone formation and resorption and the extent of radiographic change in ankylosing spondylitis. Ann. Rheum. Dis. 74, 1387–1393 (2015).
Bennett, A. N. et al. Severity of baseline magnetic resonance imaging-evident sacroiliitis and HLA-B27 status in early inflammatory back pain predict radiographically evident ankylosing spondylitis at eight years. Arthritis Rheum. 58, 3413–3418 (2008).
Huang, X.-F. et al. Genomewide association study of acute anterior uveitis identifies new susceptibility loci. Invest. Ophthalmol. Vis. Sci. 61, 3 (2020).
Thomas, G. P. et al. Genetic diagnostic profiling in axial spondyloarthritis: a real world study. Clin. Exp. Rheumatol. 35, 229–233 (2017).
Rostami, S. et al. Prediction of ankylosing spondylitis in the population-based HUNT study by a genetic risk score combining 110 single-nucleotide polymorphisms of genome-wide significance. J. Rheumatol. 47, 204–210 (2020).
Jung, S. H. et al. Developing a risk-scoring model for ankylosing spondylitis based on a combination of HLA-B27, single-nucleotide polymorphism, and copy number variant markers. J. Rheumatol. 43, 2136–2141 (2016).
Li, Z. et al. Genetic risk score prediction in ankylosing spondylitis [abstract]. Arthritis Rheumatol. 70 (Suppl. 10), 836 (2018).
Brown, M. A. et al. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann. Rheum. Dis. 55, 268–270 (1996).
Robinson, P. C., Wordsworth, B. P., Reveille, J. D. & Brown, M. A. Axial spondyloarthritis: a new disease entity, not necessarily early ankylosing spondylitis. Ann. Rheum. Dis. 72, 162–164 (2013).
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Cargill, M. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–290 (2007).
Burton, P. R. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Baeten, D. et al. Risankizumab, an IL-23 inhibitor, for ankylosing spondylitis: results of a randomised, double-blind, placebo-controlled, proof-of-concept, dose-finding phase 2 study. Ann. Rheum. Dis. 77, 1295–1302 (2018).
Poddubnyy, D., Hermann, K. G., Callhoff, J., Listing, J. & Sieper, J. Ustekinumab for the treatment of patients with active ankylosing spondylitis: results of a 28-week, prospective, open-label, proof-of-concept study (TOPAS). Ann. Rheum. Dis. 73, 817–823 (2014).
Baeten, D. et al. Anti-interleukin-17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: a randomised, double-blind, placebo-controlled trial. Lancet 382, 1705–1713 (2013).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Seager, K., Bashir, H. V., Geczy, A. F., Edmonds, J. & de Vere-Tyndall, A. Evidence for a specific B27-associated cell surface marker on lymphocytes of patients with ankylosing spondylitis. Nature 277, 68–70 (1979).
Baraliakos, X., Baerlecken, N., Witte, T., Heldmann, F. & Braun, J. High prevalence of anti-CD74 antibodies specific for the HLA class II-associated invariant chain peptide (CLIP) in patients with axial spondyloarthritis. Ann. Rheum. Dis. 73, 1079–1082 (2014).
de Winter, J. J. et al. Anti-CD74 antibodies have no diagnostic value in early axial spondyloarthritis: data from the spondyloarthritis caught early (SPACE) cohort. Arthritis Res. Ther. 20, 38 (2018).
Ranganathan, V. et al. Macrophage migration inhibitory factor induces inflammation and predicts spinal progression in ankylosing spondylitis. Arthritis Rheumatol. 69, 1796–1806 (2017).
Riechers, E. et al. Sensitivity and specificity of autoantibodies against CD74 in nonradiographic axial spondyloarthritis. Arthritis Rheumatol. 71, 729–735 (2019).
Hu, H., Du, F., Zhang, S. & Zhang, W. Serum calprotectin correlates with risk and disease severity of ankylosing spondylitis and its change during first month might predict favorable response to treatment. Mod. Rheumatol. 29, 836–842 (2019).
Klingberg, E., Carlsten, H., Hilme, E., Hedberg, M. & Forsblad-d’Elia, H. Calprotectin in ankylosing spondylitis–frequently elevated in feces, but normal in serum. Scand. J. Gastroenterol. 47, 435–444 (2012).
Klingberg, E. et al. A longitudinal study of fecal calprotectin and the development of inflammatory bowel disease in ankylosing spondylitis. Arthritis Res. Ther. 19, 21 (2017).
Cypers, H. et al. Elevated calprotectin levels reveal bowel inflammation in spondyloarthritis. Ann. Rheum. Dis. 75, 1357–1362 (2016).
Maksymowych, W. P. Biomarkers for diagnosis of axial spondyloarthritis, disease activity, prognosis, and prediction of response to therapy. Front. Immunol. 10, 305 (2019).
Liu, J. et al. Identification of disease-associated proteins by proteomic approach in ankylosing spondylitis. Biochem. Biophys. Res. Commun. 357, 531–536 (2007).
Li, T. et al. Serum disease-associated proteins of ankylosing spondylitis: results of a preliminary study by comparative proteomics. Clin. Exp. Rheumatol. 28, 201–207 (2010).
Richter, M. B. et al. The effects of intravenous pulse methylprednisolone on immunological and inflammatory processes in ankylosing spondylitis. Clin. Exp. Immunol. 53, 51–59 (1983).
Surrall, K. E., Bird, H. A. & Dixon, J. S. Caeruloplasmin, prealbumin and alpha 2-macroglobulin as potential indices of disease activity in different arthritides. Clin. Rheumatol. 6, 64–69 (1987).
Wright, C. et al. Ankylosing spondylitis monocytes show upregulation of proteins involved in inflammation and the ubiquitin proteasome pathway. Ann. Rheum. Dis. 68, 1626–1632 (2009).
Cai, A. et al. Quantitative proteomic analysis of peripheral blood mononuclear cells in ankylosing spondylitis by iTRAQ. Clin. Transl. Sci. 8, 579–583 (2015).
Fischer, R. et al. Discovery of candidate serum proteomic and metabolomic biomarkers in ankylosing spondylitis. Mol. Cell Proteom. 11, M111.013904 (2012).
Costantino, F., Breban, M. & Garchon, H. J. Genetics and functional genomics of spondyloarthritis. Front. Immunol. 9, 2933 (2018).
Fang, F., Pan, J., Xu, L., Li, G. & Wang, J. Identification of potential transcriptomic markers in developing ankylosing spondylitis: a meta-analysis of gene expression profiles. Biomed. Res. Int. 2015, 826316 (2015).
Gu, J. et al. Identification of RGS1 as a candidate biomarker for undifferentiated spondylarthritis by genome-wide expression profiling and real-time polymerase chain reaction. Arthritis Rheum. 60, 3269–3279 (2009).
Duan, R., Leo, P., Bradbury, L., Brown, M. A. & Thomas, G. P. Gene expression profiling reveals a down-regulation in immune-associated genes in AS patients. Ann. Rheum. Dis. 69, 1724–1729 (2010).
Yang, W. et al. Predisposition of six well-characterized microRNAs to syndesmophytes among Chinese patients with ankylosing spondylitis. Mod. Rheumatol. 29, 173–180 (2019).
Qian, B. P. et al. Identification of serum miR-146a and miR-155 as novel noninvasive complementary biomarkers for ankylosing spondylitis. Spine 41, 735–742 (2016).
Perez-Sanchez, C. et al. Circulating microRNAs as potential biomarkers of disease activity and structural damage in ankylosing spondylitis patients. Hum. Mol. Genet. 27, 875–890 (2018).
Assassi, S. et al. Whole-blood gene expression profiling in ankylosing spondylitis shows upregulation of Toll-like receptor 4 and 5. J. Rheumatol. 38, 87–98 (2011).
Pimentel-Santos, F. M. et al. Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue-destructive disease aspects. Arthritis Res. Ther. 13, R57 (2011).
Sharma, S. M. et al. Insights in to the pathogenesis of axial spondyloarthropathy based on gene expression profiles. Arthritis Res. Ther. 11, R168 (2009).
Gu, J. et al. A 588-gene microarray analysis of the peripheral blood mononuclear cells of spondyloarthropathy patients. Rheumatology 41, 759–766 (2002).
Smith, J. A. et al. Gene expression analysis of macrophages derived from ankylosing spondylitis patients reveals interferon-γ dysregulation. Arthritis Rheum. 58, 1640–1649 (2008).
Talpin, A. et al. Monocyte-derived dendritic cells from HLA-B27+ axial spondyloarthritis (SpA) patients display altered functional capacity and deregulated gene expression. Arthritis Res. Ther. 16, 417 (2014).
Avila Cobos, F., Vandesompele, J., Mestdagh, P. & De Preter, K. Computational deconvolution of transcriptomics data from mixed cell populations. Bioinformatics 34, 1969–1979 (2018).
Wang, X. B. et al. Transcriptome analysis of ankylosing spondylitis patients before and after TNF-alpha inhibitor therapy reveals the pathways affected. Genes. Immun. 18, 184–190 (2017).
Guma, M., Tiziani, S. & Firestein, G. S. Metabolomics in rheumatic diseases: desperately seeking biomarkers. Nat. Rev. Rheumatol. 12, 269–281 (2016).
Jiang, M. et al. Serum metabolic signatures of four types of human arthritis. J. Proteome Res. 12, 3769–3779 (2013).
Chen, R. et al. Serum fatty acid profiles and potential biomarkers of ankylosing spondylitis determined by gas chromatography-mass spectrometry and multivariate statistical analysis. Biomed. Chromatogr. 29, 604–611 (2015).
Wang, W. et al. Plasma, urine and ligament tissue metabolite profiling reveals potential biomarkers of ankylosing spondylitis using NMR-based metabolic profiles. Arthritis Res. Ther. 18, 244 (2016).
He, Z., Wang, M., Li, H. & Wen, C. GC-MS-based fecal metabolomics reveals gender-attributed fecal signatures in ankylosing spondylitis. Sci. Rep. 9, 3872 (2019).
Shao, T. et al. Characterization of ankylosing spondylitis and rheumatoid arthritis using H-1 NMR-based metabolomics of human fecal extracts. Metabolomics 12, 70 (2016).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Costello, M. E. et al. Brief report: Intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheumatol. 67, 686–691 (2015).
Wen, C. et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 18, 142 (2017).
Tito, R. Y. et al. Brief report: Dialister as a microbial marker of disease activity in spondyloarthritis. Arthritis Rheumatol. 69, 114–121 (2017).
Yin, J. et al. Shotgun metagenomics reveals an enrichment of potentially cross-reactive bacterial epitopes in ankylosing spondylitis patients, as well as the effects of TNFi therapy upon microbiome composition. Ann. Rheum. Dis. 79, 132–140 (2020).
Breban, M. et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann. Rheum. Dis. 76, 1614–1622 (2017).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Asquith, M. et al. HLA alleles associated with risk of ankylosing spondylitis and rheumatoid arthritis influence the gut microbiome. Arthritis Rheumatol. 71, 1642–1650 (2019).
Li, C. I., Samuels, D. C., Zhao, Y. Y., Shyr, Y. & Guo, Y. Power and sample size calculations for high-throughput sequencing-based experiments. Brief. Bioinform. 19, 1247–1255 (2018).
Saccenti, E. & Timmerman, M. E. Approaches to sample size determination for multivariate data: applications to PCA and PLS-DA of omics data. J. Proteome Res. 15, 2379–2393 (2016).
Li, Z. & Brown, M. A. Progress of genome-wide association studies of ankylosing spondylitis. Clin. Transl. Immunol. 6, e163 (2017).
Choi, S. W., Heng Mak, T. S. & O’Reilly, P. F. A guide to performing polygenic risk score analyses. Preprint at bioRxiv https://doi.org/10.1101/416545 (2018).
So, H. C. & Sham, P. C. Improving polygenic risk prediction from summary statistics by an empirical Bayes approach. Sci. Rep. 7, 41262 (2017).
Clemmensen, L., Witten, D., Hastie, T. & Ersbøll, B. Sparse discriminant analysis. Technometrics 53, 406–413 (2011).
Lê Cao, K.-A., Boitard, S. & Besse, P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinforma. 12, 253 (2011).
Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol. 18, 83 (2017).
Manzoni, C. et al. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 19, 286–302 (2018).
Argelaguet, R. et al. Multi-omics factor analysis — a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14, e8124 (2018).
Singh, A. et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omic assays. Bioinformatics 35, 3055–3062 (2019).
Xia, J., Gill, E. E. & Hancock, R. E. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 10, 823–844 (2015).
Holzinger, E. R., Dudek, S. M., Frase, A. T., Pendergrass, S. A. & Ritchie, M. D. ATHENA: the analysis tool for heritable and environmental network associations. Bioinformatics 30, 698–705 (2014).
Wang, W. et al. iBAG: integrative Bayesian analysis of high-dimensional multiplatform genomics data. Bioinformatics 29, 149–159 (2013).
Lee, A. H. et al. Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat. Commun. 10, 1092 (2019).
Ni, Y. & Jiang, C. Identification of potential target genes for ankylosing spondylitis treatment. Medicine 97, e9760 (2018).
Valikangas, T., Suomi, T. & Elo, L. L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 19, 1–11 (2018).
Lualdi, M. & Fasano, M. Statistical analysis of proteomics data: a review on feature selection. J. Proteom. 198, 18–26 (2019).
Aitchison, J. The Statistical Analysis of Compositional Data (Chapman & Hall, 1986).
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V. & Egozcue, J. J. Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8, 2224 (2017).
Kaul, A., Mandal, S., Davidov, O. & Peddada, S. D. Analysis of microbiome data in the presence of excess zeros. Front. Microbiol. 8, 2114 (2017).
Calle, M. L. Statistical analysis of metagenomics data. Genomics Inf. 17, e6 (2019).
Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13, R79 (2012).
Le Cao, K. A. et al. MixMC: a multivariate statistical framework to gain insight into microbial communities. PLoS One 11, e0160169 (2016).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Franzosa, E. A. et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat. Rev. Microbiol. 13, 360–372 (2015).
Oulas, A. et al. Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform. Biol. Insights. 9, 75–88 (2015).
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
McShane, L. M. & Polley, M. Y. Development of omics-based clinical tests for prognosis and therapy selection: the challenge of achieving statistical robustness and clinical utility. Clin. Trials 10, 653–665 (2013).
Vesteghem, C. et al. Implementing the FAIR Data Principles in precision oncology: review of supporting initiatives. Brief. Bioinform. 21, 936–945 (2019).
Kim, K. J. et al. Serum leptin levels are associated with the presence of syndesmophytes in male patients with ankylosing spondylitis. Clin. Rheumatol. 31, 1231–1238 (2012).
Syrbe, U. et al. Serum adipokine levels in patients with ankylosing spondylitis and their relationship to clinical parameters and radiographic spinal progression. Arthritis Rheumatol. 67, 678–685 (2015).
Genre, F. et al. Adipokines, biomarkers of endothelial activation, and metabolic syndrome in patients with ankylosing spondylitis. Biomed. Res. Int. 2014, 860651 (2014).
Hartl, A. et al. Serum levels of leptin and high molecular weight adiponectin are inversely associated with radiographic spinal progression in patients with ankylosing spondylitis: results from the ENRADAS trial. Arthritis Res. Ther. 19, 140 (2017).
Pedersen, S. J. et al. ASDAS, BASDAI and different treatment responses and their relation to biomarkers of inflammation, cartilage and bone turnover in patients with axial spondyloarthritis treated with TNFα inhibitors. Ann. Rheum. Dis. 70, 1375–1381 (2011).
de Andrade, K. R. et al. Evaluation of circulating levels of inflammatory and bone formation markers in axial spondyloarthritis. Int. Immunopharmacol. 21, 481–486 (2014).
Arends, S. et al. Higher bone turnover is related to spinal radiographic damage and low bone mineral density in ankylosing spondylitis patients with active disease: a cross-sectional analysis. PLoS One 9, e99685 (2014).
Appel, H. et al. Altered skeletal expression of sclerostin and its link to radiographic progression in ankylosing spondylitis. Arthritis Rheum. 60, 3257–3262 (2009).
Korkosz, M. et al. High disease activity in ankylosing spondylitis is associated with increased serum sclerostin level and decreased wingless protein-3a signaling but is not linked with greater structural damage. BMC Musculoskelet. Disord. 14, 99 (2013).
Turina, M. C., Yeremenko, N., Paramarta, J. E., De Rycke, L. & Baeten, D. Calprotectin (S100A8/9) as serum biomarker for clinical response in proof-of-concept trials in axial and peripheral spondyloarthritis. Arthritis Res. Ther. 16, 413 (2014).
Matzkies, F. G. et al. Markers of intestinal inflammation in patients with ankylosing spondylitis: a pilot study. Arthritis Res. Ther. 14, R261 (2012).
Schonthaler, H. B. et al. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity 39, 1171–1181 (2013).
Hammer, H. B., Kvien, T. K., Glennas, A. & Melby, K. A longitudinal study of calprotectin as an inflammatory marker in patients with reactive arthritis. Clin. Exp. Rheumatol. 13, 59–64 (1995).
Turina, M. C. et al. Calprotectin serum level is an independent marker for radiographic spinal progression in axial spondyloarthritis. Ann. Rheum. Dis. 73, 1746–1748 (2014).
Duran, A. et al. Fecal calprotectin is associated with disease activity in patients with ankylosing spondylitis. Bosn. J. Basic. Med. Sci. 16, 71–74 (2016).
Rudwaleit, M. et al. The early disease stage in axial spondylarthritis: results from the German Spondyloarthritis Inception Cohort. Arthritis Rheum. 60, 717–727 (2009).
Mattey, D. L. et al. Association of cytokine and matrix metalloproteinase profiles with disease activity and function in ankylosing spondylitis. Arthritis Res. Ther. 14, R127 (2012).
Bay-Jensen, A. C. et al. Circulating citrullinated vimentin fragments reflect disease burden in ankylosing spondylitis and have prognostic capacity for radiographic progression. Arthritis Rheum. 65, 972–980 (2013).
Bay-Jensen, A. C. et al. Circulating protein fragments of cartilage and connective tissue degradation are diagnostic and prognostic markers of rheumatoid arthritis and ankylosing spondylitis. PLoS One 8, e54504 (2013).
Taylan, A. et al. Biomarkers and cytokines of bone turnover: extensive evaluation in a cohort of patients with ankylosing spondylitis. BMC Musculoskelet. Disord. 13, 191 (2012).
Klingberg, E., Nurkkala, M., Carlsten, H. & Forsblad-d’Elia, H. Biomarkers of bone metabolism in ankylosing spondylitis in relation to osteoproliferation and osteoporosis. J. Rheumatol. 41, 1349–1356 (2014).
Kwon, S. R. et al. Dickkopf-1 level is lower in patients with ankylosing spondylitis than in healthy people and is not influenced by anti-tumor necrosis factor therapy. Rheumatol. Int. 32, 2523–2527 (2012).
Tuylu, T. et al. Fetuin-A is related to syndesmophytes in patients with ankylosing spondylitis: a case control study. Clinics 69, 688–693 (2014).
Harman, H. et al. Comparison of fetuin-A and transforming growth factor beta 1 levels in patients with spondyloarthropathies and rheumatoid arthritis. Int. J. Rheum. Dis. 20, 2020–2027 (2017).
Liu, F., Wang, F., Wang, C. C., Li, N. & Li, S. F. Expression of IL-2 and IL-11 and its significance in patients with ankylosing spondylitis. Asian Pac. J. Trop. Med. 6, 76–78 (2013).
Bal, A. et al. Comparison of serum IL-1β, sIL-2R, IL-6, and TNF-α levels with disease activity parameters in ankylosing spondylitis. Clin. Rheumatol. 26, 211–215 (2007).
Romero-Sanchez, C. et al. Association between Th-17 cytokine profile and clinical features in patients with spondyloarthritis. Clin. Exp. Rheumatol. 29, 828–834 (2011).
Gratacos, J. et al. Serum cytokines (IL-6, TNF-α, IL-1β and IFN-γ) in ankylosing spondylitis: a close correlation between serum IL-6 and disease activity and severity. Br. J. Rheumatol. 33, 927–931 (1994).
Sveaas, S. H. et al. Circulating levels of inflammatory cytokines and cytokine receptors in patients with ankylosing spondylitis: a cross-sectional comparative study. Scand. J. Rheumatol. 44, 118–124 (2015).
Mei, Y. et al. Increased serum IL-17 and IL-23 in the patient with ankylosing spondylitis. Clin. Rheumatol. 30, 269–273 (2011).
Li, G. X., Wang, S., Duan, Z. H., Zeng, Z. & Pan, F. M. Serum levels of IL-33 and its receptor ST2 are elevated in patients with ankylosing spondylitis. Scand. J. Rheumatol. 42, 226–231 (2013).
Andersen, T. et al. Increased plasma levels of IL-21 and IL-23 in spondyloarthritis are not associated with clinical and MRI findings. Rheumatol. Int. 32, 387–393 (2012).
Chen, W. S. et al. Association of serum interleukin-17 and interleukin-23 levels with disease activity in Chinese patients with ankylosing spondylitis. J. Chin. Med. Assoc. 75, 303–308 (2012).
Li, X. L. et al. Elevated serum level of IL-33 and sST2 in patients with ankylosing spondylitis: associated with disease activity and vascular endothelial growth factor. J. Investig. Med. 61, 848–851 (2013).
Ugur, M. et al. Elevated serum interleukin-23 levels in ankylosing spondylitis patients and the relationship with disease activity. Nagoya J. Med. Sci. 77, 621–627 (2015).
Han, G. W. et al. Serum levels of IL-33 is increased in patients with ankylosing spondylitis. Clin. Rheumatol. 30, 1583–1588 (2011).
Li, X. et al. Aberrant expression of microRNAs in peripheral blood mononuclear cells as candidate biomarkers in patients with axial spondyloarthritis. Int. J. Rheum. Dis. 22, 1188–1195 (2019).
Huang, J., Song, G., Yin, Z., Luo, X. & Ye, Z. Elevated miR-29a expression is not correlated with disease activity index in PBMCs of patients with ankylosing spondylitis. Mod. Rheumatol. 24, 331–334 (2014).
Keyszer, G. et al. Circulating levels of matrix metalloproteinases MMP-3 and MMP-1, tissue inhibitor of metalloproteinases 1 (TIMP-1), and MMP-1/TIMP-1 complex in rheumatic disease. Correlation with clinical activity of rheumatoid arthritis versus other surrogate markers. J. Rheumatol. 26, 251–258 (1999).
Maksymowych, W. P. et al. Serum matrix metalloproteinase 3 is an independent predictor of structural damage progression in patients with ankylosing spondylitis. Arthritis Rheum. 56, 1846–1853 (2007).
Arends, S. et al. Serum MMP-3 level as a biomarker for monitoring and predicting response to etanercept treatment in ankylosing spondylitis. J. Rheumatol. 38, 1644–1650 (2011).
Woo, J. H., Lee, H. J., Sung, I. H. & Kim, T. H. Changes of clinical response and bone biochemical markers in patients with ankylosing spondylitis taking etanercept. J. Rheumatol. 34, 1753–1759 (2007).
Chen, C. H. et al. Serum matrix metalloproteinases and tissue inhibitors of metalloproteinases in ankylosing spondylitis: MMP-3 is a reproducibly sensitive and specific biomarker of disease activity. Rheumatology 45, 414–420 (2006).
Wendling, D., Cedoz, J. P. & Racadot, E. Serum levels of MMP-3 and cathepsin K in patients with ankylosing spondylitis: effect of TNFα antagonist therapy. Jt. Bone Spine 75, 559–562 (2008).
Yang, C. et al. Serum levels of matrix metalloproteinase 3 and macrophage colony-stimulating factor 1 correlate with disease activity in ankylosing spondylitis. Arthritis Rheum. 51, 691–699 (2004).
Sun, S. et al. The active form of MMP-3 is a marker of synovial inflammation and cartilage turnover in inflammatory joint diseases. BMC Musculoskelet. Disord. 15, 93 (2014).
Gao, J. W., Zhang, K. F., Lu, J. S. & Su, T. Serum matrix metalloproteinases-3 levels in patients with ankylosing spondylitis. Genet. Mol. Res. 14, 17068–17078 (2015).
He, D. et al. Correlation of serum MMP3 and other biomarkers with clinical outcomes in patients with ankylosing spondylitis: a pilot study. Clin. Rheumatol. 36, 1819–1826 (2017).
Franck, H., Meurer, T. & Hofbauer, L. C. Evaluation of bone mineral density, hormones, biochemical markers of bone metabolism, and osteoprotegerin serum levels in patients with ankylosing spondylitis. J. Rheumatol. 31, 2236–2241 (2004).
Chen, C. H. et al. Soluble receptor activator of nuclear factor-κB ligand (RANKL) and osteoprotegerin in ankylosing spondylitis: OPG is associated with poor physical mobility and reflects systemic inflammation. Clin. Rheumatol. 29, 1155–1161 (2010).
Genre, F. et al. Osteoprotegerin correlates with disease activity and endothelial activation in non-diabetic ankylosing spondylitis patients undergoing TNF-α antagonist therapy. Clin. Exp. Rheumatol. 32, 640–646 (2014).
Visvanathan, S. et al. Effects of infliximab on markers of inflammation and bone turnover and associations with bone mineral density in patients with ankylosing spondylitis. Ann. Rheum. Dis. 68, 175–182 (2009).
Yilmaz, N. & Ozaslan, J. Biochemical bone turnover markers in patients with ankylosing spondylitis. Clin. Rheumatol. 19, 92–98 (2000).
Franck, H. & Keck, E. Serum osteocalcin and vitamin D metabolites in patients with ankylosing spondylitis. Ann. Rheum. Dis. 52, 343–346 (1993).
Saad, C. G. et al. Low sclerostin levels: a predictive marker of persistent inflammation in ankylosing spondylitis during anti-tumor necrosis factor therapy? Arthritis Res. Ther. 14, R216 (2012).
Gupta, L., Bhattacharya, S. & Aggarwal, A. Tenascin-C, a biomarker of disease activity in early ankylosing spondylitis. Clin. Rheumatol. 37, 1401–1405 (2018).
Vosse, D. et al. Association of markers of bone- and cartilage-degradation with radiological changes at baseline and after 2 years follow-up in patients with ankylosing spondylitis. Rheumatology 47, 1219–1222 (2008).
Drouart, M. et al. High serum vascular endothelial growth factor correlates with disease activity of spondylarthropathies. Clin. Exp. Immunol. 132, 158–162 (2003).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Hall, J. A., Salgado, R., Lively, T., Sweep, F. & Schuh, A. A risk-management approach for effective integration of biomarkers in clinical trials: perspectives of an NCI, NCRI, and EORTC working group. Lancet Oncol. 15, e184–e193 (2014).
Simon, R. M., Paik, S. & Hayes, D. F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl Cancer Inst. 101, 1446–1452 (2009).
Denne, J. S., Pennello, G., Zhao, L., Chang, S.-C. & Althouse, S. Identifying a subpopulation for a tailored therapy: bridging clinical efficacy from a laboratory-developed assay to a validated in vitro diagnostic test kit. Stat. Biopharmaceutical Res. 6, 78–88 (2014).
Program for the Assessment of Clinical Cancer Tests Strategy Group Members. Performance standards reporting requirements for essential assays in clinical trials. cdp.cancer.gov https://cdp.cancer.gov/scientific_programs/pacct/assay_standards.htm (2014).
Clinical and Laboratory Standards Institute. EP17-A2: Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures, 2nd edn (Clinical and Laboratory Standards Institute, 2012).
Kessler, L. G. et al. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat. Methods Med. Res. 24, 9–26 (2015).
Clinical and Laboratory Standards Institute. EP06-A: Evaluation of the Linearity of Quantitative Measurement Procedures: a Statistical Approach (Clinical and Laboratory Standards Institute, 2003).
Kimmelman, J., Resnik, D. B., Peppercorn, J. & Ratain, M. J. Burdensome research procedures in trials: why less is more. J. Natl Cancer Inst. 109, djw315 (2017).
McShane, L. M. et al. Criteria for the use of omics-based predictors in clinical trials. Nature 502, 317–320 (2013).
McShane, L. M. et al. Criteria for the use of omics-based predictors in clinical trials: explanation and elaboration. BMC Med. 11, 220 (2013).
Bossuyt, P. M. et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology 277, 826–832 (2015).
Moore, H. M. et al. Biospecimen reporting for improved study quality (BRISQ). Cancer Cytopathol. 119, 92–101 (2011).
National Cancer Institute Cancer Diagnosis Program. Templates for clinical assay development. cdp.cancer.gov https://cdp.cancer.gov/resources/templates.htm (2015).
FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource (Food and Drug Administration & National Institutes of Health, 2017).
Chau, C. H., Rixe, O., McLeod, H. & Figg, W. D. Validation of analytic methods for biomarkers used in drug development. Clin. Cancer Res. 14, 5967–5976 (2008).
Micheel, C. M. & Ball, J. R. (eds) Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease (National Academies Press, 2010).
Khan, S. R., Manialawy, Y., Wheeler, M. B. & Cox, B. J. Unbiased data analytic strategies to improve biomarker discovery in precision medicine. Drug. Discov. Today 24, 1735–1748 (2019).
Goldstein, N. S. et al. Recommendations for improved standardization of immunohistochemistry. Appl. Immunohistochem. Mol. Morphol. 15, 124–133 (2007).
Goldstein, N. S., Hunter, S., Forbes, S., Odish, E. & Tehrani, M. Estrogen receptor antibody incubation time and extent of immunoreactivity in invasive carcinoma of the breast: the importance of optimizing antibody avidity. Appl. Immunohistochem. Mol. Morphol. 15, 203–207 (2007).
Amaravadi, L. et al. 2015 White paper on recent issues in bioanalysis: focus on new technologies and biomarkers (Part 3 — LBA, biomarkers and immunogenicity). Bioanalysis 7, 3107–3124 (2015).
Arnold, M. E., Booth, B., King, L. & Ray, C. Workshop report: Crystal City VI-bioanalytical method validation for biomarkers. AAPS J. 18, 1366–1372 (2016).
Houghton, R. & Chamberlain, J. Conference report: analytical challenges in the qualification and validation of pharmacodynamic biomarkers. Bioanalysis 3, 945–948 (2011).
Lee, J. W. et al. Fit-for-purpose method development and validation for successful biomarker measurement. Pharm. Res. 23, 312–328 (2006).
Sargent, D. J., Conley, B. A., Allegra, C. & Collette, L. Clinical trial designs for predictive marker validation in cancer treatment trials. J. Clin. Oncol. 23, 2020–2027 (2005).
Spivack, S. D., Fasco, M. J., Walker, V. E. & Kaminsky, L. S. The molecular epidemiology of lung cancer. Crit. Rev. Toxicol. 27, 319–365 (1997).
Janin, M. et al. Serum 2-hydroxyglutarate production in IDH1- and IDH2-mutated de novo acute myeloid leukemia: a study by the Acute Leukemia French Association group. J. Clin. Oncol. 32, 297–305 (2014).
Berry, D. Multiplicities in cancer research: ubiquitous and necessary evils. J. Natl Cancer Inst. 104, 1124–1132 (2012).
Freemantle, N. & Calvert, M. Composite and surrogate outcomes in randomised controlled trials. BMJ 334, 756–757 (2007).
Bartley, A. N. et al. Complex patterns of altered microRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin. Cancer Res. 17, 7283–7293 (2011).
Qu, X. et al. A three-marker FISH panel detects more genetic aberrations of AR, PTEN and TMPRSS2/ERG in castration-resistant or metastatic prostate cancers than in primary prostate tumors. PLoS One 8, e74671 (2013).
Acknowledgements
The authors thank Novartis for providing assistance with the preparation of this manuscript in the form of performing a comprehensive literature survey from search terms provided by the authors, collating the reference list and optimizing figures for the Supplementary Information. K.-A.L.C. was supported in part by a National Health and Medical Research Council Career Development fellowship (GNT1159458).This work was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London and/or the NIHR Clinical Research Facility. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Author information
Authors and Affiliations
Contributions
M.A.B. and K.-A.L.C. researched data for the article and provided substantial contributions to discussions of content. All authors wrote the article and reviewed and/or edited the article before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Rheumatology thanks R. Ramonda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Australo-Anglo-American Spondyloarthritis Consortium: https://www.natureindex.com/institution-outputs/australia/australo-anglo-american-spondylitis-consortium-tasc/537afc32140ba06966000000
COSMIC: https://cancer.sanger.ac.uk/cosmic
International Genetics of Ankylosing Spondylitis Consortium: https://www.natureindex.com/institution-outputs/australia/international-genetics-of-ankylosing-spondylitis-igas-consortium/538570ad140ba07f7e000004
Supplementary information
Glossary
- Polygenic risk score
-
(PRS). A quantitative score typically involving hundreds to hundreds of thousands of genetic variants weighted by the magnitude of their association with the disease or trait of interest.
- Prior probability
-
The likelihood of an event prior to the event occurring.
- Posterior probability
-
The updated probability of an event taking into account new related information.
- Univariate and multivariate analyses
-
Analyses that take into account a single variable (univariate) or multiple variables (multivariate).
- Dependent variable
-
A variable that is being tested in an experiment that depends on the values of independent variables.
- Linear discriminant methods
-
Ways of identifying sets of quantitative variables that maximize statistical separation between sample groups (or ‘classes’).
- Partial least squares discriminant analysis
-
A multivariate regression approach to perform dimension reduction and prediction model construction.
- Dimension reduction
-
The process of reducing the number of variables under consideration by obtaining a set of linearly combined variables that carry most of the available information in the dataset.
- Overfitting
-
When a model contains more parameters than can be justified by the data and therefore might not fit additional data or extrapolate accurately.
- Matrix factorization
-
The process of decomposing a matrix into the product of two new matrices of low dimension.
- Network-based analyses
-
The analysis of experimental data on the basis of prior knowledge of interactive pathways or networks.
- Machine learning
-
A type of artificial intelligence approach whereby computational systems perform tasks on the basis of patterns and inference rather than using explicit instructions.
- Bayesian approaches
-
Probabilistic methods based on Bayes’ theorem to update probabilities for a hypothesis after obtaining new data.
Rights and permissions
About this article
Cite this article
Brown, M.A., Li, Z. & Cao, KA.L. Biomarker development for axial spondyloarthritis. Nat Rev Rheumatol 16, 448–463 (2020). https://doi.org/10.1038/s41584-020-0450-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41584-020-0450-0
This article is cited by
-
HLA-B*27 and Ankylosing Spondylitis: 50 Years of Insights and Discoveries
Current Rheumatology Reports (2023)
-
Have Therapeutics Enhanced Our Knowledge of Axial Spondyloarthritis?
Current Rheumatology Reports (2023)
-
Biomarkers in axial spondyloarthritis and low back pain: a comprehensive review
Clinical Rheumatology (2022)
-
MRI contributes to accurate and early diagnosis of non-radiographic HLA-B27 negative axial spondyloarthritis
Journal of Translational Medicine (2021)
-
MicroRNAs in Axial Spondylarthritis: an Overview of the Recent Progresses in the Field with a Focus on Ankylosing Spondylitis and Psoriatic Arthritis
Current Rheumatology Reports (2021)