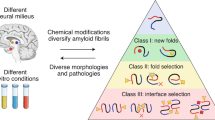
Abstract
Amyloid proteins, which are considered ‘villains’ in many neurodegenerative diseases, form enigmatic pathological strains that underlie disease pathogenesis and progression. Recent technical advances in cryogenic electron microscopy and solid-state NMR spectroscopy have enabled the high-resolution structures of full-length amyloid fibrils to be determined, initiating an era in which we have the opportunity to gain atomic-level structural understanding of pathogenic protein aggregation in neurodegenerative diseases. In this Review, we aim to explain the clinicopathological heterogeneity of neurodegenerative diseases by considering the polymorphic structures of amyloid fibrils. We decipher the structural basis for the generation of fibril polymorphs, how the fibril polymorphs differ in different disease contexts and how conformational changes alter the pathology caused by amyloid proteins during disease progression. Finally, we evaluate how this knowledge might aid clinical diagnostic and therapeutic strategies to treat neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dobson, C. M., Knowles, T. P. J. & Vendruscolo, M. The amyloid phenomenon and its significance in biology and medicine. Cold Spring Harb. Perspect. Biol. 12, a033878 (2019).
Eisenberg, D. & Jucker, M. The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012).
Peng, C., Trojanowski, J. Q. & Lee, V. M. Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 16, 199–212 (2020).
Milanesi, L. et al. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc. Natl Acad. Sci. USA 109, 20455–20460 (2012).
Engel, M. F. et al. Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc. Natl Acad. Sci. USA 105, 6033–6038 (2008).
Stancu, I. C. et al. Aggregated tau activates NLRP3-ASC inflammasome exacerbating exogenously seeded and non-exogenously seeded tau pathology in vivo. Acta Neuropathol. 137, 599–617 (2019).
Meda, L. et al. Activation of microglial cells by beta-amyloid protein and interferon-gamma. Nature 374, 647–650 (1995).
Mahul-Mellier, A. L. et al. The process of Lewy body formation, rather than simply alpha-synuclein fibrillization, is one of the major drivers of neurodegeneration. Proc. Natl Acad. Sci. USA 117, 4971–4982 (2020).
Ruan, L. et al. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446 (2017).
Olzscha, H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 (2011).
Suzuki, G. et al. α-synuclein strains that cause distinct pathologies differentially inhibit proteasome. Elife 9, e56825 (2020).
Winslow, A. R. et al. α-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J. Cell Biol. 190, 1023–1037 (2010).
Cuervo, A. M., Stefanis, L., Fredenburg, R., Lansbury, P. T. & Sulzer, D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 (2004).
Prusiner, S. B. Biology and genetics of prions causing neurodegeneration. Annu. Rev. Genet. 47, 601–623 (2013).
Jucker, M. & Walker, L. C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 (2013).
Luk, K. C. et al. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953 (2012). This study reports that intrastriatal inoculation of α-syn fibrils causes cell-to-cell transmission and PD-like Lewy pathology in mouse brain. The PD mouse model developed in this study has been widely used for investigating the pathological properties of α-syn fibrils.
Prusiner, S. B. et al. Evidence for alpha-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl Acad. Sci. USA 112, E5308–E5317 (2015).
Kaufman, S. K. et al. Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron 92, 796–812 (2016).
Kovacs, G. G. et al. Distribution patterns of tau pathology in progressive supranuclear palsy. Acta Neuropathol. 140, 99–119 (2020).
Kim, S. et al. Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron 103, 627–641 (2019).
Guo, J. L. et al. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell 154, 103–117 (2013).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Braak, H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211 (2003).
Vogel, J. W. et al. Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nat. Med. 27, 871–881 (2021).
Geddes, A. J., Parker, K. D., Atkins, E. D. & Beighton, E. “Cross-beta” conformation in proteins. J. Mol. Biol. 32, 343–358 (1968).
Jahn, T. R. et al. The common architecture of cross-beta amyloid. J. Mol. Biol. 395, 717–727 (2010).
Astbury, W. T., Dickinson, S. & Bailey, K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 29, 2351–2360.1 (1935).
Qiang, W., Yau, W. M., Lu, J. X., Collinge, J. & Tycko, R. Structural variation in amyloid-beta fibrils from Alzheimer’s disease clinical subtypes. Nature 541, 217–221 (2017).
Falcon, B. et al. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561, 137–140 (2018).
Gremer, L. et al. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science 358, 116–119 (2017).
Fitzpatrick, A. W. P. et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017). This study reveals, for the first time, the cryo-EM structures of tau amyloid fibrils directly extracted from the brains of patients with AD.
Li, Y. et al. Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy. Cell Res. 28, 897–903 (2018).
Tuttle, M. D. et al. Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 (2016).
Paravastu, A. K., Leapman, R. D., Yau, W. M. & Tycko, R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc. Natl Acad. Sci. USA 105, 18349–18354 (2008).
Walti, M. A. et al. Atomic-resolution structure of a disease-relevant Abeta(1-42) amyloid fibril. Proc. Natl Acad. Sci. USA 113, E4976–E4984 (2016).
Radamaker, L. et al. Cryo-EM reveals structural breaks in a patient-derived amyloid fibril from systemic AL amyloidosis. Nat. Commun. 12, 875 (2021).
Shankar, G. M. et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 14, 837–842 (2008).
Kayed, R. et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T. & Lansbury, P. T. Jr. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418, 291 (2002).
Winner, B. et al. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc. Natl Acad. Sci. USA 108, 4194–4199 (2011).
Otzen, D. & Riek, R. Functional amyloids. Cold Spring Harb. Perspect. Biol. 11, a033860 (2019).
Sawaya, M. R., Hughes, M. P., Rodriguez, J. A., Riek, R. & Eisenberg, D. S. The expanding amyloid family: structure, stability, function, and pathogenesis. Cell 184, 4857–4873 (2021).
Wei, G. et al. Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. Chem. Soc. Rev. 46, 4661–4708 (2017).
Hardy, J. A. & Higgins, G. A. Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185 (1992).
Crowther, R. A. Straight and paired helical filaments in Alzheimer disease have a common structural unit. Proc. Natl Acad. Sci. USA 88, 2288–2292 (1991).
Kidd, M. Paired helical filaments in electron microscopy of Alzheimer’s disease. Nature 197, 192–193 (1963).
Spillantini, M. G. et al. Alpha-synuclein in Lewy bodies. Nature 388, 839–840 (1997).
Polymeropoulos, M. H. et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997).
Lashuel, H. A., Overk, C. R., Oueslati, A. & Masliah, E. The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 14, 38–48 (2013).
Wetzel, R., Shivaprasad, S. & Williams, A. D. Plasticity of amyloid fibrils. Biochemistry 46, 1–10 (2007).
Benson, M. D. et al. Amyloid nomenclature 2018: recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 25, 215–219 (2018).
Sunde, M. et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273, 729–739 (1997).
Nelson, R. et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 (2005). This study demonstrates the high structural diversity of amyloid-like fibril spines formed in vitro by amyloidogenic peptides at the atomic level.
Khurana, R. et al. Mechanism of thioflavin T binding to amyloid fibrils. J. Struct. Biol. 151, 229–238 (2005).
Collinge, J., Sidle, K. C., Meads, J., Ironside, J. & Hill, A. F. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383, 685–690 (1996).
Zhao, K. et al. Parkinson’s disease-related phosphorylation at Tyr39 rearranges alpha-synuclein amyloid fibril structure revealed by cryo-EM. Proc. Natl Acad. Sci. USA 117, 20305–20315 (2020). This study reveals the structural basis by which a PD-related phosphorylation determines the formation of a new fold within an α-syn fibril with enhanced neurotoxicity.
Zhao, K. et al. Parkinson’s disease associated mutation E46K of alpha-synuclein triggers the formation of a distinct fibril structure. Nat. Commun. 11, 2643 (2020).
Wang, L. Q. et al. Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nat. Struct. Mol. Biol. 27, 598–602 (2020).
Sun, Y. et al. Cryo-EM structure of full-length alpha-synuclein amyloid fibril with Parkinson’s disease familial A53T mutation. Cell Res. 30, 360–362 (2020).
Guerrero-Ferreira, R. et al. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. Elife 8, e48907 (2019).
Petkova, A. T. et al. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science 307, 262–265 (2005). This study reveals that Aβ can form amyloid fibrils with distinct molecular structures and neurotoxicity.
Petkova, A. T., Yau, W. M. & Tycko, R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry 45, 498–512 (2006).
Colletier, J. P. et al. Molecular basis for amyloid-beta polymorphism. Proc. Natl Acad. Sci. USA 108, 16938–16943 (2011).
Sawaya, M. R. et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007).
Li, B. et al. Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609 (2018).
Guerrero-Ferreira, R. et al. Cryo-EM structure of alpha-synuclein fibrils. Elife 7, e36402 (2018).
Shi, Y. et al. Structure-based classification of tauopathies. Nature 598, 359–363 (2021). This study reports the structure of tau fibrils extracted from brain tissues of patients with progressive supranuclear palsy, and demonstrates that different tau fibril structures can be used for hierarchical classification of tauopathies.
Zhang, W. et al. Novel tau filament fold in corticobasal degeneration. Nature 580, 283–287 (2020).
Falcon, B. et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 (2019).
Schweighauser, M. et al. Structures of alpha-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020). This study reports, for the first time, the cryo-EM structures of α-syn fibrils extracted from patients’ brains, and shows the structural difference between in vitro-prepared α-syn fibrils and brain-extracted α-syn fibrils.
Boyer, D. R. et al. The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc. Natl Acad. Sci. USA 117, 3592–3602 (2020).
Fersht, A. R. From the first protein structures to our current knowledge of protein folding: delights and scepticisms. Nat. Rev. Mol. Cell Biol. 9, 650–654 (2008).
Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997).
Dill, K. A. & MacCallum, J. L. The protein-folding problem, 50 years on. Science 338, 1042–1046 (2012).
Tayeb-Fligelman, E. et al. The cytotoxic Staphylococcus aureus PSMalpha3 reveals a cross-alpha amyloid-like fibril. Science 355, 831–833 (2017).
Luo, F. et al. Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol. 25, 341–346 (2018).
Hughes, M. P. et al. Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science 359, 698–701 (2018).
Kant, R. et al. A structural analysis of amyloid polymorphism in disease: clues for selective vulnerability? Preprint at https://doi.org/10.1101/2021.03.01.433317 (2021).
McKinley, M. P., Bolton, D. C. & Prusiner, S. B. A protease-resistant protein is a structural component of the scrapie prion. Cell 35, 57–62 (1983).
Safar, J., Roller, P. P., Gajdusek, D. C. & Gibbs, C. J. Jr. Thermal stability and conformational transitions of scrapie amyloid (prion) protein correlate with infectivity. Protein Sci. 2, 2206–2216 (1993).
Li, D. et al. Designed amyloid fibers as materials for selective carbon dioxide capture. Proc. Natl Acad. Sci. USA 111, 191–196 (2014).
Torrent, J. et al. High pressure induces scrapie-like prion protein misfolding and amyloid fibril formation. Biochemistry 43, 7162–7170 (2004).
Boyer, D. R. et al. Structures of fibrils formed by alpha-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol. 26, 1044–1052 (2019).
Arakhamia, T. et al. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 180, 633–644 e612 (2020). This study reports the cryo-EM structure of tau fibrils extracted from the brains of patients with corticobasal degeneration, and demonstrates that PTMs are important in determining the structural polymorphs of tau fibril in different tauopathies.
Li, D. & Liu, C. Hierarchical chemical determination of amyloid polymorphs in neurodegenerative disease. Nat. Chem. Biol. 17, 237–245 (2021).
Miura, T., Suzuki, K., Kohata, N. & Takeuchi, H. Metal binding modes of Alzheimer’s amyloid beta-peptide in insoluble aggregates and soluble complexes. Biochemistry 39, 7024–7031 (2000).
Zarranz, J. J. et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164–173 (2004).
Zhang, W. et al. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. Elife 8, e43584 (2019).
Lovestam, S. et al. Seeded assembly in vitro does not replicate the structures of alpha-synuclein filaments from multiple system atrophy. FEBS Open Bio 11, 999–1013 (2021).
Fan, Y. et al. Different structures and pathologies of alpha-synuclein fibrils derived from preclinical and postmortem patients of Parkinson’s disease. Preprint at https://doi.org/10.1101/2021.11.02.467019 (2021).
Peng, C. et al. Cellular milieu imparts distinct pathological alpha-synuclein strains in alpha-synucleinopathies. Nature 557, 558–563 (2018). This study shows that different intracellular milieus of neurons and oligodendrocytes can induce different α-syn fibril strains with distinct pathology.
Sanders, D. W. et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 (2014).
Mao, X. et al. Pathological alpha-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 353, aah3374 (2016).
Goedert, M. Alzheimer’s and Parkinson’s diseases: the prion concept in relation to assembled Abeta, tau, and alpha-synuclein. Science 349, 1255555 (2015).
Zhang, S. et al. Mechanistic basis for receptor-mediated pathological alpha-synuclein fibril cell-to-cell transmission in Parkinson’s disease. Proc. Natl Acad. Sci. USA 118, e2011196118 (2021).
Davis, A. A., Leyns, C. E. G. & Holtzman, D. M. Intercellular spread of protein aggregates in neurodegenerative disease. Annu. Rev. Cell Dev. Biol. 34, 545–568 (2018).
Fares, M. B., Jagannath, S. & Lashuel, H. A. Reverse engineering Lewy bodies: how far have we come and how far can we go? Nat. Rev. Neurosci. 22, 111–131 (2021).
Shahnawaz, M. et al. Discriminating alpha-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 578, 273–277 (2020).
Strohaker, T. et al. Structural heterogeneity of alpha-synuclein fibrils amplified from patient brain extracts. Nat. Commun. 10, 5535 (2019).
Long, H. et al. Wild-type alpha-synuclein inherits the structure and exacerbated neuropathology of E46K mutant fibril strain by cross-seeding. Proc. Natl Acad. Sci. USA 118, e2012435118 (2021).
Brahmachari, S. et al. Activation of tyrosine kinase c-Abl contributes to alpha-synuclein-induced neurodegeneration. J. Clin. Invest. 126, 2970–2988 (2016).
McGlinchey, R. P. & Lee, J. C. Cysteine cathepsins are essential in lysosomal degradation of alpha-synuclein. Proc. Natl Acad. Sci. USA 112, 9322–9327 (2015).
McGlinchey, R. P. et al. C-terminal alpha-synuclein truncations are linked to cysteine cathepsin activity in Parkinson’s disease. J. Biol. Chem. 294, 9973–9984 (2019).
Moors, T. E. et al. The subcellular arrangement of alpha-synuclein proteoforms in the Parkinson’s disease brain as revealed by multicolor STED microscopy. Acta Neuropathol. 142, 423–448 (2021).
Sorrentino, Z. A. et al. Physiological C-terminal truncation of alpha-synuclein potentiates the prion-like formation of pathological inclusions. J. Biol. Chem. 293, 18914–18932 (2018).
Luk, K. C. et al. Molecular and Biological Compatibility with Host Alpha-Synuclein Influences Fibril Pathogenicity. Cell Rep. 16, 3373–3387 (2016).
Kollmer, M. et al. Cryo-EM structure and polymorphism of Abeta amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 10, 4760 (2019).
Lu, J. X. et al. Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 154, 1257–1268 (2013). This study shows that Aβ40 fibrils seeded from patients with AD with distinct clinical histories exhibit different molecular architectures.
Ghosh, U., Thurber, K. R., Yau, W. M. & Tycko, R. Molecular structure of a prevalent amyloid-beta fibril polymorph from Alzheimer’s disease brain tissue. Proc. Natl Acad. Sci. USA 118, e2023089118 (2021).
Yang, Y. et al. Cryo-EM structures of amyloid-beta 42 filaments from human brains. Science 375, 167–172 (2022).
Shi, Y. et al. Cryo-EM structures of tau filaments from Alzheimer’s disease with PET ligand APN-1607. Acta Neuropathol. 141, 697–708 (2021).
Burger, D. et al. Cryo-EM structure of alpha-synuclein fibrils amplified by PMCA from PD and MSA patient brains. Preprint at https://doi.org/10.1101/2021.07.08.451588 (2021).
Lövestam, S. et al. Assembly of recombinant tau into filaments identical to those of Alzheimer’s disease and chronic traumatic encephalopathy. Elife 11, e76494 (2022).
Schütz, A. et al. Atomic-resolution three-dimensional structure of amyloid beta fibrils bearing the Osaka mutation. Angew. Chem. Int. Ed. 54, 331–335 (2015).
Wesseling, H. et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer’s disease. Cell 183, 1699–1713 (2020). This study demonstrates that tau proteins isolated from the brain tissues of patients with AD exhibit different PTM patterns at different stages of AD.
Morris, M. et al. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat. Neurosci. 18, 1183–1189 (2015).
Dujardin, S. et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 26, 1256–1263 (2020).
Glynn, C. et al. Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core. Nat. Struct. Mol. Biol. 27, 417–423 (2020).
Aoyagi, A. et al. Abeta and tau prion-like activities decline with longevity in Alzheimer’s disease brains. Sci. Transl. Med. 11, eaat8462 (2019).
Lucic, V., Rigort, A. & Baumeister, W. Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol. 202, 407–419 (2013).
Trinkaus, V. A. et al. In situ architecture of neuronal alpha-synuclein inclusions. Nat. Commun. 12, 2110 (2021).
Bauerlein, F. J. B., Fernandez-Busnadiego, R. & Baumeister, W. Investigating the structure of neurotoxic protein aggregates inside cells. Trends Cell Biol. 30, 951–966 (2020).
Irwin, D. J. et al. Deep clinical and neuropathological phenotyping of Pick disease. Ann. Neurol. 79, 272–287 (2016).
Montenigro, P. H., Corp, D. T., Stein, T. D., Cantu, R. C. & Stern, R. A. Chronic traumatic encephalopathy: historical origins and current perspective. Annu. Rev. Clin. Psychol. 11, 309–330 (2015).
Crary, J. F. et al. Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol. 128, 755–766 (2014).
Forrest, S. L., Kril, J. J. & Halliday, G. M. Cellular and regional vulnerability in frontotemporal tauopathies. Acta Neuropathol. 138, 705–727 (2019).
Gotz, J., Halliday, G. & Nisbet, R. M. Molecular pathogenesis of the tauopathies. Annu. Rev. Pathol. 14, 239–261 (2019).
Saito, Y. et al. Staging of argyrophilic grains: an age-associated tauopathy. J. Neuropathol. Exp. Neurol. 63, 911–918 (2004).
Botez, G., Probst, A., Ipsen, S. & Tolnay, M. Astrocytes expressing hyperphosphorylated tau protein without glial fibrillary tangles in argyrophilic grain disease. Acta Neuropathol. 98, 251–256 (1999).
Kovacs, G. G. et al. Aging-related tau astrogliopathy (ARTAG): harmonized evaluation strategy. Acta Neuropathol. 131, 87–102 (2016).
Braak, H., Sastre, M. & Del Tredici, K. Development of alpha-synuclein immunoreactive astrocytes in the forebrain parallels stages of intraneuronal pathology in sporadic Parkinson’s disease. Acta Neuropathol. 114, 231–241 (2007).
Wakabayashi, K., Hayashi, S., Yoshimoto, M., Kudo, H. & Takahashi, H. NACP/alpha-synuclein-positive filamentous inclusions in astrocytes and oligodendrocytes of Parkinson’s disease brains. Acta Neuropathol. 99, 14–20 (2000).
Halliday, G. M., Holton, J. L., Revesz, T. & Dickson, D. W. Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. 122, 187–204 (2011).
Terada, S. et al. Glial involvement in diffuse Lewy body disease. Acta Neuropathol. 105, 163–169 (2003).
Jellinger, K. A. Lewy body/alpha-synucleinopathy in schizophrenia and depression: a preliminary neuropathological study. Acta Neuropathol. 117, 423–427 (2009).
Jellinger, K. A. Lewy body-related alpha-synucleinopathy in the aged human brain. J. Neural Transm. 111, 1219–1235 (2004).
Shishido, T. et al. alpha-synuclein accumulation in skin nerve fibers revealed by skin biopsy in pure autonomic failure. Neurology 74, 608–610 (2010).
Kaufmann, H., Hague, K. & Perl, D. Accumulation of alpha-synuclein in autonomic nerves in pure autonomic failure. Neurology 56, 980–981 (2001).
Coon, E. A., Singer, W. & Low, P. A. Pure autonomic failure. Mayo Clin. Proc. 94, 2087–2098 (2019).
Kovari, E., Burkhardt, K., Lobrinus, J. A. & Bouras, C. Lewy body dysphagia. Acta Neuropathol. 114, 295–298 (2007).
Jackson, M., Lennox, G., Balsitis, M. & Lowe, J. Lewy body dysphagia. J. Neurol. Neurosurg. Psychiatry 58, 756–758 (1995).
Nakamura, K. et al. Accumulation of phosphorylated alpha-synuclein in subpial and periventricular astrocytes in multiple system atrophy of long duration. Neuropathology 36, 157–167 (2016).
Vabulas, M. et al. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2, a004390 (2010).
Acknowledgements
The authors thank Y. Ma, Y. Fan and H. Long for assisting with the preparation of figures and tables. They also thank the National Natural Science Foundation of China (grants 82188101, 32170683, 31872716 and 32171236), the Major State Basic Research Development Program (grant 2019YFE0120600), the Science and Technology Commission of Shanghai Municipality (grants 20XD1425000 and 2019SHZDZX02), the Chinese Academy of Science Project for Young Scientists in Basic Research (grant YSBR-009) and the Shanghai Pilot Program for Basic Research–Chinese Academy of Science, Shanghai Branch (grant CYJ-SHFY-2022-005), for funding support.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
RCSB Protein Data Bank: https://www.rcsb.org/
Glossary
- Amyloid fibrils
-
Insoluble aggregates, formed by repetitive supermolecular assembly of a misfolded protein, in which the protein units are ordered in β-strand-rich structures.
- Self-propagation
-
Process in which preformed amyloid fibrils of a protein nucleate the conversion of the soluble form of the protein into the amyloid form to generate more fibrils.
- Cross-β structure
-
A common structural feature of amyloid fibrils that arises from β-sheet bundles in which the direction of β-strands is nearly perpendicular to the fibril axis.
- Cryogenic electron microscopy
-
(cryo-EM). A transmission electron microscopy technique that is applied to samples that have been rapidly frozen into a glass-like state.
- Solid-state NMR spectroscopy
-
An NMR technology used to investigate the chemical structure and dynamics of solids and semi-solids at an atomic level.
- Protein strains
-
Proteins that form the unique fibril structures that underlie the pathological presentation of a disease.
- Conformational strains
-
Forms of a protein with distinct fibril structures.
- Prion diseases
-
A family of neurodegenerative disorders caused by misfolded prion protein that affect both humans and animals.
- Isoforms
-
Different forms of a protein produced from alternative splicing of the same gene or from similar genes.
- Fibril polymorphs
-
Amyloid fibrils with different structures formed from the same protein.
- Post-translational modifications
-
(PTMs). Biochemical modifications of one or more amino acids within a protein that occur after translation.
- Protein misfolding
-
A common cellular event in which a protein fails to achieve its native structure, resulting in functional abnormality.
- Intrinsically disordered proteins
-
Proteins that lack unique 3D structures but may transit to ordered physiological structures upon interaction with binding partners or to pathological structures under disease conditions.
- α-Helix
-
A type of secondary structure in which the protein chain is coiled as a result of regularly spaced hydrogen bonding between residues and in which the side chains are left reaching outwards.
- β-Strand
-
A type of secondary structure in which the protein chain is extended as a result of the backbone forming hydrogen bonds with the adjacent chains and in which the side chains alternate on both sides.
- Free energy
-
Also called Gibbs free energy. A thermodynamic quantity that expresses the amount of work that can be done by a system. In protein folding, the change of free energy is used to describe the thermodynamic stability of a protein.
- Energy landscape
-
A model that describes protein folding as a funnel-like landscape gradually biased towards the energetically optimal structure.
- Cofactors
-
Substances essential for the construction of the fibril.
- Charged pockets
-
Cavities on the surface or in the interior of a fibril that contain charged residues.
- Salt bridges
-
Electrostatic attractions between oppositely charged amino acid side chains.
- β-Hairpin motif
-
A protein structural motif in which the adjacent antiparallel β-strands are connected either by backbone hydrogen bonding (as usually seen in native structures) or by side chain interactions (as seen in fibril structures).
- Seeds
-
Preformed amyloid fibrils that can serve as a nucleus to template and accelerate the conversion of soluble proteins to fibrils.
Rights and permissions
About this article
Cite this article
Li, D., Liu, C. Conformational strains of pathogenic amyloid proteins in neurodegenerative diseases. Nat Rev Neurosci 23, 523–534 (2022). https://doi.org/10.1038/s41583-022-00603-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41583-022-00603-7
This article is cited by
-
Synthetic β-sheets mimicking fibrillar and oligomeric structures for evaluation of spectral X-ray scattering technique for biomarker quantification
Cell & Bioscience (2024)
-
The involvement of α-synucleinopathy in the disruption of microglial homeostasis contributes to the pathogenesis of Parkinson’s disease
Cell Communication and Signaling (2024)
-
Glymphatic inhibition exacerbates tau propagation in an Alzheimer’s disease model
Alzheimer's Research & Therapy (2024)
-
Multifunctional nanoparticle-mediated combining therapy for human diseases
Signal Transduction and Targeted Therapy (2024)
-
Cryo-EM structures reveal variant Tau amyloid fibrils between the rTg4510 mouse model and sporadic human tauopathies
Cell Discovery (2024)