Abstract
The pathology of Alzheimer disease (AD) damages structural and functional brain networks, resulting in cognitive impairment. The results of recent connectomics studies have now linked changes in structural and functional network organization in AD to the patterns of amyloid-β and tau accumulation and spread, providing insights into the neurobiological mechanisms of the disease. In addition, the detection of gene-related connectome changes might aid in the early diagnosis of AD and facilitate the development of personalized therapeutic strategies that are effective at earlier stages of the disease spectrum. In this article, we review studies of the associations between connectome changes and amyloid-β and tau pathologies as well as molecular genetics in different subtypes and stages of AD. We also highlight the utility of connectome-derived computational models for replicating empirical findings and for tracking and predicting the progression of biomarker-indicated AD pathophysiology.
Key points
-
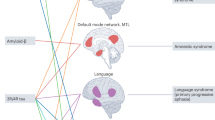
Amyloid-β (Aβ) pathology is associated with decreased hub connectivity in the default-mode network (DMN) during the preclinical stage of Alzheimer disease (AD) and the association extends to other brain networks as the disease progresses.
-
Selective hub vulnerability might explain the preferential accumulation of Aβ in the medial hubs of the DMN, and of tau in medial temporal lobe hubs, in preclinical AD.
-
Tau pathology spreads from the medial temporal lobe hubs — along structural connections — to other brain regions, supporting the pathogenic spread hypothesis.
-
Aβ pathology has a common role in driving DMN hypo-connectivity in late-onset AD, autosomal-dominant AD and early-onset AD; however, the association between Aβ pathology and DMN hypoconnectivity is regulated by different genetic variants across AD subtypes.
-
Spatial gene expression profiles might contribute to the relationships between the patterns of Aβ and tau accumulation and patterns of structural and functional connectome changes in AD.
-
Computational modelling studies will be important for understanding the role of the connectome in relation to progression of Aβ, tau and other pathogenic features of AD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
11 August 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41582-021-00554-0
References
Squire, L. R., Stark, C. E. L. & Clark, R. E. The medial temporal lobe. Annu. Rev. Neurosci. 27, 279–306 (2004).
Dubois, B. et al. Clinical diagnosis of Alzheimer’s disease: recommendations of the International Working Group. Lancet Neurol. 4422, 1–13 (2021).
Sperling, R. A. et al. Amyloid deposition is associated with impaired default network function in older persons without dementia. Neuron 63, 178–188 (2009).
Palmqvist, S. et al. Earliest accumulation of β-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat. Commun. 8, 1214 (2017). This article provides compelling evidence that Aβ accumulation preferentially starts in the DMN.
Buckner, R. L. & DiNicola, L. M. The brain’s default network: updated anatomy, physiology and evolving insights. Nat. Rev. Neurosci. 20, 593–608 (2019).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Small, S. A., Schobel, S. A., Buxton, R. B., Witter, M. P. & Barnes, C. A. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 12, 585–601 (2011).
Hampel, H. et al. Blood-based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat. Rev. Neurol. 14, 639–652 (2018).
Jack, C. R. et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 12, 207–216 (2013).
Jack, C. R. & Holtzman, D. M. Biomarker modeling of Alzheimer’s disease. Neuron 80, 1347–1358 (2013).
Jack, C. R., Hampel, H. J., Universities, S., Cu, M. & Petersen, R. C. A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 87, 539–547 (2016).
Jack, C. R. et al. Prevalence of biologically vs clinically defined Alzheimer spectrum entities using the National Institute on Aging-Alzheimer’s Association Research Framework. JAMA Neurol. 76, 1174–1183 (2019).
Gaiteri, C., Mostafavi, S., Honey, C. J., De Jager, P. L. & Bennett, D. A. Genetic variants in Alzheimer disease-molecular and brain network approaches. Nat. Rev. Neurol. 12, 413–427 (2016).
Bateman, R. J. et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 367, 795–804 (2012).
Bateman, R. J. et al. Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res. Ther. 3, 1 (2011).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Sims, R., Hill, M. & Williams, J. The multiplex model of the genetics of Alzheimer’s disease. Nat. Neurosci. 38, 30–34 (2020).
Verghese, P. B., Castellano, J. M. & Holtzman, D. M. Apolipoprotein E in Alzheimer’s disease and other neurological disorders. Lancet Neurol. 10, 241–252 (2011).
Yamazaki, Y., Zhao, N., Caulfield, T. R., Liu, C.-C. & Bu, G. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 15, 501–518 (2019).
Masters, C. L. et al. Alzheimer’s disease. Nat. Rev. Dis. Prim. 1, 15056 (2015).
Gallagher, M. & Koh, M. T. Episodic memory on the path to Alzheimer’s disease. Curr. Opin. Neurobiol. 21, 929–934 (2011).
Ossenkoppele, R. et al. Amyloid burden and metabolic function in early-onset Alzheimer’s disease: parietal lobe involvement. Brain 135, 2115–2125 (2012).
Cho, H. et al. Amyloid deposition in early onset versus late onset Alzheimer’s disease. J. Alzheimers Dis. 35, 813–821 (2013).
Balasa, M. et al. Clinical features and APOE genotype of pathologically proven early-onset Alzheimer disease. Neurology 76, 1720–1725 (2011).
Snowden, J. S. et al. The clinical diagnosis of early-onset dementias: diagnostic accuracy and clinicopathological relationships. Brain 134, 2478–2492 (2011).
Gordon, B. A. et al. Tau PET in autosomal dominant Alzheimer’ s disease: relationship with cognition, dementia and other biomarkers. Brain 142, 1063–1076 (2019).
Bullmore, E. & Sporns, O. The economy of brain network organization. Nat. Rev. Neurosci. 13, 336–349 (2012).
Park, H. J. & Friston, K. Structural and functional brain networks: from connections to cognition. Science 342, 1238411 (2013).
Stam, C. J. Modern network science of neurological disorders. Nat. Rev. Neurosci. 15, 683–695 (2014). This review article provides a comprehensive review of the application of graph theory and network science to multiple brain disorders.
Fornito, A., Zalesky, A. & Breakspear, M. The connectomics of brain disorders. Nat. Rev. Neurosci. 16, 159–172 (2015).
Menon, V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn. Sci. 15, 483–506 (2011).
Palop, J. J. & Mucke, L. Network abnormalities and interneuron dysfunction in Alzheimer disease. Nat. Rev. Neurosci. 17, 777–792 (2016).
Delbeuck, X., Van der Linden, M. & Collette, F. Alzheimer’s disease as a disconnection syndrome. Neuropsychol. Rev. 13, 79–92 (2003).
Catani, M. & Ffytche, D. H. The rises and falls of disconnection syndromes. Brain 128, 2224–2239 (2005).
Sporns, O. Contributions and challenges for network models in cognitive neuroscience. Nat. Neurosci. 17, 652–660 (2014).
Bassett, D. S. & Sporns, O. Network neuroscience. Nat. Neurosci. 20, 353–364 (2017).
Yu, M. Benchmarking metrics for inferring functional connectivity from multi-channel EEG and MEG: a simulation study. Chaos 30, 123124 (2020).
Yu, M., Hillebrand, A., Gouw, A. A. & Stam, C. J. Horizontal visibility graph transfer entropy (HVG-TE): a novel metric to characterize directed connectivity in large-scale brain networks. Neuroimage 156, 249–264 (2017).
Hillebrand, A. et al. Direction of information flow in large-scale resting-state networks is frequency-dependent. Proc. Natl Acad. Sci. USA 113, 3867–3872 (2016).
Bullmore, E. & Sporns, O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 312–312 (2009). This review article provides a thorough review of structural and functional connectome studies.
Rubinov, M. & Sporns, O. Weight-conserving characterization of complex functional brain networks. Neuroimage 56, 2068–2079 (2011).
Rubinov, M. & Sporns, O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069 (2010).
Sporns, O., Tononi, G. & Kötter, R. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, 0245–0251 (2005).
Smith, S. M. et al. Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 17, 666–682 (2013).
Bassett, D. S. & Bullmore, E. T. Small-world brain networks revisited. Neuroscientist 23, 499–516 (2016).
van den Heuvel, M. P. & Sporns, O. Network hubs in the human brain. Trends Cogn. Sci. 17, 683–696 (2013).
van den Heuvel, M. P. & Sporns, O. Rich-club organization of the human connectome. J. Neurosci. 31, 15775–15786 (2011).
Sporns, O. & Betzel, R. F. Modular brain networks. Annu. Rev. Psychol. 67, 613–640 (2016).
Meunier, D., Lambiotte, R. & Bullmore, E. T. Modular and hierarchically modular organization of brain networks. Front. Neurosci. 4, 200 (2010).
Sporns, O. Network attributes for segregation and integration in the human brain. Curr. Opin. Neurobiol. 23, 162–171 (2013).
Betzel, R. F. & Bassett, D. S. Multi-scale brain networks. Neuroimage 160, 73–83 (2016).
Yu, M. et al. Selective impairment of hippocampus and posterior hub areas in Alzheimer’s disease: an MEG-based multiplex network study. Brain 140, 1466–1485 (2017). This paper was the first to describe relationships between MEG-based functional multiplex network topology and Aβ and tau pathologies as well as cognitive decline in AD.
Power, J. D., Schlaggar, B. L., Lessov-Schlaggar, C. N. & Petersen, S. E. Evidence for hubs in human functional brain networks. Neuron 79, 798–813 (2013).
Zuo, X. N. et al. Network centrality in the human functional connectome. Cereb. Cortex 22, 1862–1875 (2012).
Gong, G. et al. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex 19, 524–536 (2009).
Hagmann, P. et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 6, 1479–1493 (2008).
Mišić, B., Goñi, J., Betzel, R. F., Sporns, O. & McIntosh, A. R. A network convergence zone in the hippocampus. PLoS Comput. Biol. 10, e1003982 (2014).
Battaglia, F. P., Benchenane, K., Sirota, A., Pennartz, C. M. A. & Wiener, S. I. The hippocampus: Hub of brain network communication for memory. Trends Cogn. Sci. 15, 310–318 (2011).
Swanson, L. W., Hahn, J. D. & Sporns, O. Organizing principles for the cerebral cortex network of commissural and association connections. Proc. Natl Acad. Sci. USA 114, E9692–E9701 (2017).
Tononi, G., Sporns, O. & Edelman, G. M. A measure for brain complexity: relating functional segregation and integration in the nervous system. Proc. Natl Acad. Sci. USA 91, 5033–5037 (1994).
Yeo, B. T. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011).
Schaefer, A. et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex 28, 3095–3114 (2018).
Cole, M. W. et al. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat. Neurosci. 16, 1348–1355 (2013).
Cole, M. W., Bassett, D. S., Power, J. D., Braver, T. S. & Petersen, S. E. Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251 (2014).
Smith, S. et al. Structural variability in the human brain reflects fine-grained functional architecture at the population level. J. Neurosci. 39, 6136–6149 (2019).
Fox, M. D. & Raichle, M. E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 8, 700–711 (2007).
Fulcher, B. D. & Fornito, A. A transcriptional signature of hub connectivity in the mouse connectome. Proc. Natl Acad. Sci. USA 113, 1435–1440 (2016).
Arnatkevicˇiu¯te∙, A., Fulcher, B. D. & Fornito, A. Uncovering the transcriptional correlates of hub connectivity in neural networks. Front. Neural Circuits 13, 47 (2019).
Thompson, P. M., Ge, T., Glahn, D. C., Jahanshad, N. & Nichols, T. E. Genetics of the connectome. Neuroimage 80, 475–488 (2013).
Fornito, A., Arnatkevicˇiu¯te∙, A. & Fulcher, B. D. Bridging the gap between connectome and transcriptome. Trends Cogn. Sci. 23, 34–50 (2019). This article reviews the relationships between brain connectome topology and brain-wide gene expression.
De Haan, W. et al. Disrupted modular brain dynamics reflect cognitive dysfunction in Alzheimer’s disease. Neuroimage 59, 3085–3093 (2012).
Yu, M. et al. Hierarchical clustering in minimum spanning trees. Chaos 25, 023107 (2015).
Yu, M. et al. Different functional connectivity and network topology in behavioral variant of frontotemporal dementia and Alzheimer’s disease: an EEG study. Neurobiol. Aging 42, 150–162 (2016).
Buckner, R. L. et al. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J. Neurosci. 29, 1860–1873 (2009).
Crossley, N. A. et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 137, 2382–2395 (2014).
Filippi, M. et al. Changes in functional and structural brain connectome along the Alzheimer’s disease continuum. Mol. Psychiatry 25, 230–239 (2020).
Schöll, M. et al. PET imaging of tau deposition in the aging human brain. Neuron 89, 971–982 (2016).
Bassett, D. S., Zurn, P. & Gold, J. I. On the nature and use of models in network neuroscience. Nat. Rev. Neurosci. 19, 566–578 (2018).
Pievani, M., Filippini, N., Van Den Heuvel, M. P., Cappa, S. F. & Frisoni, G. B. Brain connectivity in neurodegenerative diseases - From phenotype to proteinopathy. Nat. Rev. Neurol. 10, 620–633 (2014).
Tijms, B. M. et al. Alzheimer’s disease: connecting findings from graph theoretical studies of brain networks. Neurobiol. Aging 34, 2023–2036 (2013).
Alexander-Bloch, A., Giedd, J. N. & Bullmore, E. Imaging structural co-variance between human brain regions. Nat. Rev. Neurosci. 14, 322–336 (2013).
Tijms, B. M., Seris, P., Willshaw, D. J. & Lawrie, S. M. Similarity-based extraction of individual networks from gray matter MRI scans. Cereb. Cortex 22, 1530–1541 (2012).
Tijms, B. M. et al. Single-subject grey matter graphs in Alzheimer’s disease. PLoS One 8, e58921 (2013).
Tijms, B. M. et al. Gray matter network disruptions and amyloid beta in cognitively normal adults. Neurobiol. Aging 37, 154–160 (2016).
ten Kate, M. et al. Gray matter network disruptions and regional amyloid beta in cognitively normal adults. Front. Aging Neurosci. 10, 67 (2018).
Tijms, B. M. et al. Gray matter networks and clinical progression in subjects with predementia Alzheimer’s disease. Neurobiol. Aging 61, 75–81 (2018).
Dicks, E., van der Flier, W. M., Scheltens, P., Barkhof, F. & Tijms, B. M. Single-subject grey matter networks predict future cortical atrophy in preclinical Alzheimer’s disease. Neurobiol. Aging 94, 71–80 (2020).
Voevodskaya, O. et al. Altered structural network organization in cognitively normal individuals with amyloid pathology. Neurobiol. Aging 64, 15–24 (2018).
Prescott, J. W. et al. The Alzheimer structural connectome: changes in cortical network topology with increased amyloid plaque burden. Radiology 273, 175–184 (2014).
Jonkman, L. et al. Relationship between β-amyloid and structural network topology in decedents without dementia. Neurology 95, e532–e544 (2020). This article was the first to describe relationships between Aβ accumulation and structural brain network topology in decedents without dementia.
Mito, R. et al. Fibre-specific white matter reductions in Alzheimer’s disease and mild cognitive impairment. Brain 141, 888–902 (2018).
Kantarci, K. et al. White matter integrity determined with diffusion tensor imaging in older adults without dementia: Influence of amyloid load and neurodegeneration. JAMA Neurol. 71, 1547–1554 (2014).
Rabin, J. S. et al. Global white matter diffusion characteristics predict longitudinal cognitive change independently of amyloid status in clinically normal older adults. Cereb. Cortex 29, 1251–1262 (2019).
Parra, M. A. et al. Memory binding and white matter integrity in familial Alzheimer’s disease. Brain 138, 1355–1369 (2015).
Kantarci, K. et al. White-matter integrity on DTI and the pathologic staging of Alzheimer’s disease. Neurobiol. Aging 56, 172–179 (2017).
Iturria-Medina, Y. & Evans, A. C. On the central role of brain connectivity in neurodegenerative disease progression. Front. Aging Neurosci. 7, 90 (2015).
Kuang, W., Cieslak, M., Greene, C., Grafton, S. T. & Carlson, J. M. Sensitivity analysis of human brain structural network construction Kuang. Netw. Neurosci. 1, 446–467 (2017).
Powell, F., Tosun, D., Sadeghi, R., Weiner, M. & Raj, A. Preserved structural network organization mediates pathology spread in Alzheimer’s disease spectrum despite loss of white matter tract integrity. J. Alzheimers Dis. 65, 747–764 (2018).
Maier-Hein, K. H. et al. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 8, 1349 (2017).
Thomas, C. et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc. Natl Acad. Sci. USA 111, 16574–16579 (2014).
Schilling, K. G. et al. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage 185, 1–11 (2019).
Millar, P. R. et al. Evaluating resting-state BOLD variability in relation to biomarkers of preclinical Alzheimer disease. Neurobiol. Aging 96, 233–245 (2020).
Hedden, T. et al. Disruption of functional connectivity in clinically normal older adults harboring amyloid burden. J. Neurosci. 29, 12686–12694 (2009).
Johnson, K. A., Sperling, R. A. & Sepulcre, J. Functional connectivity in Alzheimer’s disease: measurement and meaning. Biol. Psychiatry 74, 318–319 (2013).
Koch, K. et al. Disrupted intrinsic networks link amyloid-β pathology and impaired cognition in prodromal Alzheimer’s disease. Cereb. Cortex 25, 4678–4688 (2015).
Drzezga, A. et al. Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain 134, 1635–1646 (2011).
Lehmann, M. et al. Intrinsic connectivity networks in healthy subjects explain clinical variability in Alzheimer’s disease. Proc. Natl Acad. Sci. USA 110, 11606–11611 (2013).
Jones, D. T. et al. Cascading network failure across the Alzheimer’s disease spectrum. Brain 139, 547–562 (2016).
Schultz, A. P. et al. Longitudinal degradation of the default/salience network axis in symptomatic individuals with elevated amyloid burden. NeuroImage Clin. 26, 102052 (2020).
Berron, D. et al. Medial temporal lobe connectivity and its associations with cognition in early Alzheimer’s disease. Brain 143, 1233–1248 (2020).
Ossenkoppele, R. & Hansson, O. Towards clinical application of tau PET tracers for diagnosing dementia due to Alzheimer’s disease. Alzheimers Dement. https://doi.org/10.1002/alz.12356 (2021).
Jacobs, H. I. L. et al. Structural tract alterations predict downstream tau accumulation in amyloid-positive older individuals. Nat. Neurosci. 21, 424–431 (2018). This article provides evidence that tau spreads through structural connections facilitated by Aβ pathology.
Shigemoto, Y. et al. Association of deposition of tau and amyloid-β proteins with structural connectivity changes in cognitively normal older adults and Alzheimer’s disease spectrum patients. Brain Behav. 8, e01145 (2018).
Harrison, T. M. et al. Longitudinal tau accumulation and atrophy in aging and alzheimer disease. Ann. Neurol. 85, 229–240 (2019).
Joie, R. L. et al. Prospective longitudinal atrophy in Alzheimer’ s disease correlates with the intensity and topography of baseline tau-PET. Sci. Transl. Med. 12, eaau5732 (2020).
Reimand, J., Collij, L., Scheltens, P., Femke Bouwman & Ossenkoppele, R. Amyloid-β CSF/PET discordance vs tau load 5 years later: it takes two to tangle. Neurology 95, e2648–e2657 (2020).
Mattsson, N. et al. Predicting diagnosis and cognition with 18 F-AV-1451 tau PET and structural MRI in Alzheimer’s disease. Alzheimers Dement. 15, 570–580 (2019).
Jacobs, H. I. L. et al. The presubiculum links incipient amyloid and tau pathology to memory function in older persons. Neurology 94, e1916–e1928 (2020).
Iaccarino, L. et al. Spatial relationships between molecular pathology and neurodegeneration in the Alzheimer’s disease continuum. Cereb. Cortex 31, 1–14 (2020).
Wang, L. et al. Cerebrospinal fluid Aβ42, phosphorylated tau181, and resting-state functional connectivity. JAMA Neurol. 70, 1242–1248 (2013).
Canuet, L. et al. Network disruption and cerebrospinal fluid amyloid-beta and phospho-tau levels in mild cognitive impairment. J. Neurosci. 35, 10325–10330 (2015).
Schultz, A. P. et al. Phases of hyperconnectivity and hypoconnectivity in the default mode and salience networks track with amyloid and tau in clinically normal individuals. J. Neurosci. 37, 4323–4331 (2017).
Maass, A. et al. Alzheimer’s pathology targets distinct memory networks in the ageing brain. Brain 142, 2492–2509 (2019). This study reports evidence that tau and Aβ pathologies target distinctive functional brain networks in the ageing brain.
Harrison, T. M. et al. Tau deposition is associated with functional isolation of the hippocampus in aging. Nat. Commun. 10, 4900 (2019).
Gordon, B. A. et al. Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer’s disease: a longitudinal study. Lancet Neurol. 17, 211–212 (2018).
Benzinger, T. L. S. et al. Regional variability of imaging biomarkers in autosomal dominant Alzheimer’s disease. Proc. Natl Acad. Sci. USA 110, E4502–E4509 (2013).
Hansson, O. et al. Tau pathology distribution in Alzheimer’s disease corresponds differentially to cognition-relevant functional brain networks. Front. Neurosci. 11, 167 (2017).
Franzmeier, N. et al. Functional brain architecture is associated with the rate of tau accumulation in Alzheimer’s disease. Nat. Commun. 11, 347 (2020). This study reports the spatial relationships between that tau accumulation and functional brain networks in AD.
Franzmeier, N. et al. Functional connectivity associated with tau levels in ageing, Alzheimer’s, and small vessel disease. Brain 142, 1093–1107 (2019).
Ossenkoppele, R. et al. Tau covariance patterns in Alzheimer’s disease patients match intrinsic connectivity networks in the healthy brain. NeuroImage Clin. 23, 101848 (2019).
Cope, T. E. et al. Tau burden and the functional connectome in Alzheimer’s disease and progressive supranuclear palsy. Brain 141, 550–567 (2018). This study was the first to assess the relationship between tau burden and fMRI-based functional network topology characterized by graph-theoretic measures.
Schöll, M. et al. Biomarkers for tau pathology. Mol. Cell. Neurosci. 97, 18–33 (2019).
Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci. 19, 687–700 (2018). This article provides a comprehensive review of neuroimaging studies in different types and stages of AD.
Pereira, J. B. et al. Amyloid and tau accumulate across distinct spatial networks and are differentially associated with brain connectivity. eLife 8, e50830 (2019).
Sepulcre, J. et al. Tau and amyloid β proteins distinctively associate to functional network changes in the aging brain. Alzheimers Dement. 13, 1261–1269 (2017).
Huijbers, W. et al. Tau accumulation in clinically normal older adults is associated with increases in hippocampal fMRI activity. J. Neurosci. 39, 548–556 (2019).
Adams, J. N., Maass, A., Harrison, T. M., Baker, S. L. & Jagust, W. J. Cortical tau deposition follows patterns of entorhinal functional connectivity in aging. eLife 8, e49132 (2019).
Therriault, J. et al. Association of apolipoprotein e ε4 with medial temporal tau Independent of amyloid-β. JAMA Neurol. 77, 470–479 (2020).
Therriault, J. et al. APOEε4 potentiates the relationship between amyloid-β and tau pathologies. Mol. Psychiatry https://doi.org/10.1038/s41380-020-0688-6 (2020).
Wolk, D. A. & Dickerson, B. C. Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer’s disease. Proc. Natl Acad. Sci. USA 107, 10256–10261 (2010).
Chiesa, P. A., Cavedo, E., Lista, S., Thompson, P. M. & Hampel, H. Revolution of resting-state functional neuroimaging genetics in Alzheimer’s disease. Trends Neurosci. 40, 469–480 (2017).
Sheline, Y. I. et al. APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF A 42. J. Neurosci. 30, 17035–17040 (2010).
Chiesa, P. A. et al. Differential default mode network trajectories in asymptomatic individuals at risk for Alzheimer’s disease. Alzheimers Dement. 15, 940–950 (2019). This article provides evidence that APOE ε4 leads to changes in DMN, independent of Aβ pathology.
Wang, J. et al. Apolipoprotein E ε4 modulates functional brain connectome in Alzheimer’s disease. Hum. Brain Mapp. 36, 1828–1846 (2015).
Contreras, J. A. et al. Functional connectivity among brain regions affected in Alzheimer’s disease is associated with CSF TNF-α in APOE4 carriers. Neurobiol. Aging 86, 112–122 (2020).
Machulda, M. M. et al. Effect of APOE ε4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch. Neurol. 68, 1131–1136 (2011).
Butt, O. H. et al. Network dysfunction in cognitively normal APOE ε4 carriers is related to subclinical tau. Alzheimers Dement. https://doi.org/10.1002/alz.12375 (2021).
Meije Wink, A. et al. Functional brain network centrality is related to APOE genotype in cognitively normal elderly. Brain Behav. 8, e01080 (2018).
Wang, L. et al. Alzheimer disease family history impacts resting state functional connectivity. Ann. Neurol. 72, 571–577 (2012).
Verfaillie, S. C. J. et al. Subjective cognitive decline is associated with altered default mode network connectivity in individuals with a family history of Alzheimer’s disease. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 3, 463–472 (2018).
Vogel, J. W. et al. Brain properties predict proximity to symptom onset in sporadic Alzheimer’s disease. Brain 141, 1871–1883 (2018).
Brown, J. A. et al. Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proc. Natl Acad. Sci. USA 108, 20760–20765 (2011).
Shu, N. et al. Effects of APOE promoter polymorphism on the topological organization of brain structural connectome in nondemented elderly. Hum. Brain Mapp. 36, 4847–4858 (2015).
Chang, P. et al. The effects of an APOE promoter polymorphism on human white matter connectivity during non-demented aging. J. Alzheimers Dis. 55, 77–87 (2016).
Ma, C. et al. Disrupted brain structural connectivity: pathological interactions between genetic APOE ε4 status and developed MCI condition. Mol. Neurobiol. 54, 6999–7007 (2017).
Chen, Y. et al. Disrupted functional and structural networks in cognitively normal elderly subjects with the APOE ε4 allele. Neuropsychopharmacology 40, 1181–1191 (2015).
Korthauer, L. E., Zhan, L., Ajilore, O., Leow, A. & Driscoll, I. Disrupted topology of the resting state structural connectome in middle-aged APOE ε4 carriers. Neuroimage 178, 295–305 (2018). This study shows that APOE ε4-related structural connectome changes occur even in middle-aged individuals.
Elsheikh, S. S. M., Chimusa, E. R., Mulder, N. J. & Crimi, A. Genome-wide association study of brain connectivity changes for Alzheimer’s disease. Sci. Rep. 10, 1433 (2020).
Jahanshad, N. et al. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc. Natl Acad. Sci. USA 110, 4768–4773 (2013).
Feinstein, Y. et al. F-spondin and mindin: two structurally and functionally related genes expressed in the hippocampus that promote outgrowth of embryonic hippocampal neurons. Development 126, 3637–3648 (1999).
Hoe, H. S. & William Rebeck, G. Functional interactions of APP with the apoE receptor family. J. Neurochem. 106, 2263–2271 (2008).
Hafez, D. M. et al. F-spondin gene transfer improves memory performance and reduces amyloid-β levels in mice. Neuroscience 223, 465–472 (2012).
Lee, S. et al. White matter hyperintensities are a core feature of Alzheimer’s disease: Evidence from the dominantly inherited Alzheimer network. Ann. Neurol. 79, 929–939 (2016).
Araque Caballero, M. Á. et al. White matter diffusion alterations precede symptom onset in autosomal dominant Alzheimer’s disease. Brain 141, 3065–3080 (2018).
Vermunt, L. et al. Single-subject grey matter network trajectories over the disease course of autosomal dominant Alzheimer disease. Brain Commun. 2, fcaa102 (2020). This article reports evidence that single-subject structural grey matter covariance network metrics can track the progression of autosomal-dominant AD.
Chhatwal, J. P. et al. Impaired default network functional connectivity in autosomal dominant Alzheimer disease. Neurology 81, 736–744 (2013).
Thomas, J. B. et al. Functional connectivity in autosomal dominant and late-onset Alzheimer disease. JAMA Neurol. 71, 1111–1122 (2014).
Chhatwal, J. P. et al. Preferential degradation of cognitive networks differentiates Alzheimer’s disease from ageing. Brain 141, 1486–1500 (2018).
Franzmeier, N. et al. Left frontal hub connectivity delays cognitive impairment in autosomal-dominant and sporadic Alzheimer’s disease. Brain 141, 1186–1200 (2018).
Mendez, M. F. Early-onset Alzheimer disease and its variants. Contin. Lifelong Learn. Neurol. 25, 34–51 (2019).
Mendez, M. F. Early-onset Alzheimer disease. Neurol. Clin. 35, 263–281 (2017).
Filippi, M. et al. Brain network connectivity differs in early-onset neurodegenerative dementia. Neurology 89, 1764–1772 (2017). A functional brain network study shows distinctive network connectivity patterns in early-onset AD and frontotemporal dementia.
Lee, E.-S. et al. Default mode network functional connectivity in early and late mild cognitive impairment. Alzheimer Dis. Assoc. Disord. 30, 289–296 (2016).
Buckner, R. L., Andrews-Hanna, J. R. & Schacter, D. L. The brain’s default network: Anatomy, function, and relevance to disease. Ann. N. Y. Acad. Sci. 1124, 1–38 (2008).
Li, K. C. et al. Distinct patterns of interhemispheric connectivity in patients with early- and late-onset Alzheimer’s disease. Front. Aging Neurosci. 10, 261 (2018).
Daianu, M. et al. An advanced white matter tract analysis in frontotemporal dementia and early-onset Alzheimer’s disease. Brain Imaging Behav. 10, 1038–1053 (2016).
Daianu, M. et al. Disrupted rich club network in behavioral variant frontotemporal dementia and early-onset Alzheimer’s disease. Hum. Brain Mapp. 37, 868–883 (2016).
Goedert, M. Alzheimer’s and Parkinson’s diseases: the prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 349, 61–69 (2015).
Frost, B. & Diamond, M. I. Prion-like mechanisms in neurodegenerative diseases. Nat. Rev. Neurosci. 11, 155–159 (2010).
Fornari, S., Schäfer, A., Jucker, M., Goriely, A. & Kuhl, E. Prion-like spreading of Alzheimer’s disease within the brain’s connectome. J. R. Soc. Interface 16, 20190356 (2019).
Peng, C., Trojanowski, J. Q. & Lee, V. M. Y. Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 16, 199–212 (2020).
He, Z. et al. Amyloid-β plaques enhance Alzheimer’s brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat. Med. 24, 29–38 (2018).
Mezias, C., LoCastro, E., Xia, C. & Raj, A. Connectivity, not region-intrinsic properties, predicts regional vulnerability to progressive tau pathology in mouse models of disease. Acta Neuropathol. Commun. 5, 61 (2017).
Walsh, D. M. & Selkoe, D. J. A critical appraisal of the pathogenic protein spread hypothesis of neurodegeneration. Nat. Rev. Neurosci. 17, 251–260 (2016).
Small, S. A. & Swanson, L. W. A network explanation of Alzheimer’s regional vulnerability. Cold Spring Harb. Symp. Quant. Biol. 83, 193–200 (2018).
Roussarie, J. P. et al. Selective neuronal vulnerability in Alzheimer’s disease: a network-based analysis. Neuron 107, 821–835 (2020).
Jagust, W. Vulnerable neural systems and the borderland of brain aging and neurodegeneration. Neuron 77, 219–234 (2013).
Mattsson, N., Schott, J. M., Hardy, J., Turner, M. R. & Zetterberg, H. Selective vulnerability in neurodegeneration: Insights from clinical variants of Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 87, 1000–1004 (2016).
Pascoal, T. A. et al. Aβ-induced vulnerability propagates via the brain’s default mode network. Nat. Commun. 10, 2353 (2019).
Vaishnavi, S. N. et al. Regional aerobic glycolysis in the human brain. Proc. Natl Acad. Sci. USA 107, 17757–17762 (2010).
Tomasi, D. & Volkow, N. D. Association between functional connectivity hubs and brain networks. Cereb. Cortex 21, 2003–2013 (2011).
Vlassenko, A. G. et al. Spatial correlation between brain aerobic glycolysis and amyloid-β (Aβ) deposition. Proc. Natl Acad. Sci. USA 107, 17763–17767 (2010).
Hanseeuw, B. J. et al. Fluorodeoxyglucose metabolism associated with tau-amyloid interaction predicts memory decline. Ann. Neurol. 81, 583–596 (2017).
Adams, J. N., Lockhart, S. N., Li, L. & Jagust, W. J. Relationships between tau and glucose metabolism reflect Alzheimer’s disease pathology in cognitively normal older adults. Cereb. Cortex 29, 1997–2009 (2019).
Ossenkoppele, R. et al. Tau PET patterns mirror clinical and neuroanatomical variability in Alzheimer’s disease. Brain 139, 1551–1567 (2016).
de Haan, W., Mott, K., van Straaten, E. C. W., Scheltens, P. & Stam, C. J. Activity dependent degeneration explains hub vulnerability in Alzheimer’s disease. PLoS Comput. Biol. 8, e1002582 (2012).
Jafari, Z., Kolb, B. E. & Mohajerani, M. H. Neural oscillations and brain stimulation in Alzheimer’s disease. Prog. Neurobiol. 194, 101878 (2020).
de Haan, W., van Straaten, E. C. W., Gouw, A. A. & Stam, C. J. Altering neuronal excitability to preserve network connectivity in a computational model of Alzheimer’s disease. PLoS Comput. Biol. 13, e1005707 (2017).
Chételat, G. et al. Amyloid-PET and 18F-FDG-PET in the diagnostic investigation of Alzheimer’s disease and other dementias. Lancet Neurol. 19, 951–962 (2020).
Altmann, A., Ng, B., Landau, S. M., Jagust, W. J. & Greicius, M. D. Regional brain hypometabolism is unrelated to regional amyloid plaque burden. Brain 138, 3734–3746 (2015).
Vogel, J. W. et al. Spread of pathological tau proteins through communicating neurons in human Alzheimer’s disease. Nat. Commun. 11, 2612 (2020). This computational modelling study provides evidence in humans that tau spreads through neuronal network pathways facilitated by Aβ accumulation.
Detrez, J. R. et al. Progressive tau aggregation does not alter functional brain network connectivity in seeded hTau.P301L mice. Neurobiol. Dis. 143, 105011 (2020).
Vogel, J. W. et al. Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nat. Med. 27, 871–881 (2021).
Raj, A. & Powell, F. Models of network spread and network degeneration in brain disorders. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 3, 788–797 (2018).
Torok, J., Maia, P. D., Powell, F., Pandya, S. & Raj, A. A method for inferring regional origins of neurodegeneration. Brain 141, 863–876 (2018).
Acosta, D., Powell, F., Zhao, Y. & Raj, A. Regional vulnerability in Alzheimer’s: the role of cell-autonomous and transneuronal processes. Alzheimers Dement. 14, 797–810 (2018).
Raj, A., Kuceyeski, A. & Weiner, M. A network diffusion model of disease progression in dementia. Neuron 73, 1204–1215 (2012).
Stam, C. J. & Reijneveld, J. C. Graph theoretical analysis of complex networks in the brain. Nonlinear Biomed. Phys. 1, 3 (2007).
van der Kant, R., Goldstein, L. S. B. & Ossenkoppele, R. Amyloid-β-independent regulators of tau pathology in Alzheimer disease. Nat. Rev. Neurosci. 21, 21–35 (2020).
Hawrylycz, M. et al. Canonical genetic signatures of the adult human brain. Nat. Neurosci. 18, 1832–1844 (2015). This landmark paper identified reproducible gene expression signatures related to functional brain connectivity.
Grothe, M. J. et al. Molecular properties underlying regional vulnerability to Alzheimer’s disease pathology. Brain 141, 2755–2771 (2018).
Sepulcre, J. et al. Neurogenetic contributions to amyloid beta and tau spreading in the human cortex. Nat. Med. 24, 1910–1918 (2018). This article was one of the first linking Aβ accumulation and tau spreading with brain-wide gene expression in the human AD.
Rauch, J. N. et al. LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385 (2020).
Shinohara, M., Tachibana, M., Kanekiyo, T. & Bu, G. Role of LRP1 in the pathogenesis of Alzheimer’s disease: Evidence from clinical and preclinical studies. J. Lipid Res. 58, 1267–1281 (2017).
Tachibana, M. et al. APOE4-mediated amyloid-β pathology depends on its neuronal receptor LRP1. J. Clin. Invest. 129, 1272–1277 (2019).
Liu, Q. et al. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron 56, 66–78 (2007).
Tsvetanov, K. A., Henson, R. N. A. & Rowe, J. B. Separating vascular and neuronal effects of age on fMRI BOLD signals. Phil. Trans. R. Soc. B 376, 20190631 (2021).
Purkayastha, S. et al. Impaired cerebrovascular hemodynamics are associated with cerebral white matter damage. J. Cereb. Blood Flow. Metab. 34, 228–234 (2014).
Buckley, R. F. et al. Associations between baseline amyloid, sex, and APOE on subsequent tau accumulation in cerebrospinal fluid. Neurobiol. Aging 78, 178–185 (2019).
Sintini, I. et al. Longitudinal neuroimaging biomarkers differ across Alzheimer’s disease phenotypes. Brain 143, 2281–2294 (2020).
Myszczynska, M. A. et al. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat. Rev. Neurol. 16, 440–456 (2020).
McIntosh, A. R. & Mišic´, B. Multivariate statistical analyses for neuroimaging data. Annu. Rev. Psychol. 64, 499–525 (2013).
Smith, S. M. et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat. Neurosci. 18, 1565–1567 (2015).
Yu, M. et al. Childhood trauma history is linked to abnormal brain connectivity in major depression. Proc. Natl Acad. Sci. USA 116, 8582–8590 (2019).
Yu, M. et al. Structural brain measures linked to clinical phenotypes in major depression replicate across clinical centres. Mol. Psychiatry https://doi.org/10.1038/s41380-021-01039-8 (2021).
Ferreira, D., Nordberg, A. & Westman, E. Biological subtypes of Alzheimer disease: a systematic review and meta-analysis. Neurology 94, 436–448 (2020).
Long, J. M. & Holtzman, D. M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179, 312–339 (2019).
Yu, M. et al. Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Hum. Brain Mapp. 39, 4213–4227 (2018). This paper was one of the first to use a statistical harmonization technique, called ComBat, to eliminate the impact of site effects on functional connectivity and brain network measures.
Badhwar, A. et al. A multiomics approach to heterogeneity in Alzheimer’s disease: focused review and roadmap. Brain 143, 1315–1331 (2020).
Nativio, R. et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 52, 1024–1035 (2020).
Power, J. D. et al. Functional network organization of the human brain. Neuron 72, 655–678 (2011).
Johnson, K. A., Fox, N. C., Sperling, R. A. & Klunk, W. E. Brain imaging in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2, a006213 (2012).
Hansson, O. Biomarkers for neurodegenerative diseases. Nat. Med. 27, 954–963 (2021).
Dubois, B. et al. Preclinical Alzheimer’s disease: definition, natural history, and diagnostic criteria. Alzheimers Dement. 12, 292–323 (2016).
Sperling, R. A. et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 280–292 (2011).
Jessen, F. et al. The characterisation of subjective cognitive decline. Lancet Neurol. 19, 271–278 (2020).
Gauthier, S. et al. Mild cognitive impairment. Lancet 367, 1262–1270 (2006).
Jack, C. R. et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 14, 535–562 (2018).
Ebenau, J. L. et al. ATN classification and clinical progression in subjective cognitive decline. Neurology 95, e46–e58 (2020).
Mattsson-Carlgren, N. et al. The implications of different approaches to define AT(N) in Alzheimer disease. Neurology 94, e2233–e2244 (2020).
Cousins, K. A. Q. et al. ATN status in amnestic and non-amnestic Alzheimer’s disease and frontotemporal lobar degeneration. Brain 143, 2295–2311 (2020).
Badji, A. & Westman, E. Cerebrovascular pathology in Alzheimer’s disease: hopes and gaps. Psychiatry Res. Neuroimaging 306, 111184 (2020).
Love, S. & Miners, J. S. Cerebrovascular disease in ageing and Alzheimer’s disease. Acta Neuropathol. 131, 645–658 (2016).
Newman, M. E. J. Communities, modules and large-scale structure in networks. Nat. Phys. 8, 25–31 (2011).
Fortunato, S. & Hric, D. Community detection in networks: a user guide. Phys. Rep. 659, 1–44 (2016).
Newman, M. E. J. J. & Girvan, M. Finding and evaluating community structure in networks. Phys. Rev. E 69, 026113 (2004).
Blondel, V. D., Guillaume, J.-L. L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, P10008 (2008).
Fortunato, S. & Barthélemy, M. Resolution limit in community detection. Proc. Natl Acad. Sci. USA 104, 36–41 (2007).
Guimera, R. & Amaral, L. A. N. Functional cartography of complex metabolic networks. Nature 433, 895–900 (2005).
Stam, C. J. et al. The trees and the forest: characterization of complex brain networks with minimum spanning trees. Int. J. Psychophysiol. 92, 129–138 (2014).
Zalesky, A., Fornito, A. & Bullmore, E. On the use of correlation as a measure of network connectivity. Neuroimage 60, 2096–2106 (2012).
van Wijk, B. C. M., Stam, C. J. & Daffertshofer, A. Comparing brain networks of different size and connectivity density using graph theory. PLoS One 5, e13701 (2010).
Fornito, A., Zalesky, A. & Breakspear, M. Graph analysis of the human connectome: Promise, progress, and pitfalls. Neuroimage 80, 426–444 (2013).
Garrison, K. A., Scheinost, D., Finn, E. S., Shen, X. & Constable, R. T. The (in)stability of functional brain network measures across thresholds. Neuroimage 118, 651–661 (2015).
de Reus, M. A. & van den Heuvel, M. P. Estimating false positives and negatives in brain networks. Neuroimage 70, 402–409 (2013).
van den Heuvel, M. P. et al. Proportional thresholding in resting-state fMRI functional connectivity networks and consequences for patient-control connectome studies: Issues and recommendations. Neuroimage 152, 437–449 (2017).
Tewarie, P., van Dellen, E., Hillebrand, A. & Stam, C. J. The minimum spanning tree: an unbiased method for brain network analysis. Neuroimage 104, 177–188 (2015).
Roberts, J. A., Perry, A., Roberts, G., Mitchell, P. B. & Breakspear, M. Consistency-based thresholding of the human connectome. Neuroimage 145, 118–129 (2017).
Drakesmith, M. et al. Overcoming the effects of false positives and threshold bias in graph theoretical analyses of neuroimaging data. Neuroimage 118, 313–333 (2015).
Bielczyk, N. Z. et al. Thresholding functional connectomes by means of mixture modeling. Neuroimage 171, 402–414 (2018).
Braun, U. et al. Test-retest reliability of resting-state connectivity network characteristics using fMRI and graph theoretical measures. Neuroimage 59, 1404–1412 (2012).
Brettschneider, J., Del Tredici, K., Lee, V. M. Y. & Trojanowski, J. Q. Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nat. Rev. Neurosci. 16, 109–120 (2015).
Scearce-Levie, K., Sanchez, P. E. & Lewcock, J. W. Leveraging preclinical models for the development of Alzheimer disease therapeutics. Nat. Rev. Drug Discov. 19, 447–462 (2020).
Bero, A. W. et al. Neuronal activity regulates the regional vulnerability to amyloid-beta deposition. Nat. Neurosci. 14, 750–756 (2011).
Wu, J. W. et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 19, 1085–1092 (2016).
Iba, M. et al. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J. Neurosci. 33, 1024–1037 (2013).
Mudher, A. et al. What is the evidence that tau pathology spreads through prion-like propagation? Acta Neuropathol. Commun. 5, 99 (2017).
Jucker, M. & Walker, L. C. Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat. Neurosci. 21, 1341–1349 (2018).
Stancu, I. C. et al. Templated misfolding of Tau by prion-like seeding along neuronal connections impairs neuronal network function and associated behavioral outcomes in Tau transgenic mice. Acta Neuropathol. 129, 875–894 (2015).
Bassil, F. et al. Amyloid-beta (Aβ) plaques promote seeding and spreading of alpha-synuclein and tau in a mouse model of Lewy body disorders with Aβ pathology. Neuron 105, 260–275.e6 (2020).
Götz, J., Bodea, L. G. & Goedert, M. Rodent models for Alzheimer disease. Nat. Rev. Neurosci. 19, 583–598 (2018).
Kitazawa, M., Medeiros, R. & LaFerla, M. F. Transgenic mouse models of Alzheimer disease: developing a better model as a tool for therapeutic interventions. Curr. Pharm. Des. 18, 1131–1147 (2012).
Myers, A. & McGonigle, P. Overview of transgenic mouse models for Alzheimer’s disease. Curr. Protoc. Neurosci. 89, e81 (2019).
Breakspear, M. Dynamic models of large-scale brain activity. Nat. Neurosci. 20, 340–352 (2017). This review discusses multiple computational models of brain activity dynamics.
Cabral, J., Kringelbach, M. L. & Deco, G. Exploring the network dynamics underlying brain activity during rest. Prog. Neurobiol. 114, 102–131 (2014).
Iturria-Medina, Y., Sotero, R. C., Toussaint, P. J. & Evans, A. C. Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders. PLoS Comput. Biol. 10, e1003956 (2014).
Acknowledgements
The preparation of this manuscript was supported in part by U.S. National Institutes of Health grants: R01 AG197711, P30 AG10133, 1U01AG024904, R01 CA129769, R01 AG057739, R01 LM013463, R01 AG068193 and U01 AG068057. The authors would like to thank Dr. Martijn van den Heuvel for assisting with the drawing of Fig. 1b. The authors thank Dr. Kwangsik Nho and Dr. Shannon Risacher for valuable discussions.
Author information
Authors and Affiliations
Contributions
M. Y. researched data for the article, made a substantial contribution to discussion of content, wrote the article, and reviewed and edited the manuscript before submission. O. S. and A. J. S. researched data for the article, made a substantial contribution to discussion of content, and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Neurology thanks B. Bendlin, who co-reviewed with A. Kohli, A. Raj and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Allen Human Brain Atlas: http://human.brain-map.org/
Alzheimer’s Disease Neuroimaging Initiative: http://adni.loni.usc.edu/
BioFINDER: https://biofinder.se/
Human Connectome Project: https://www.humanconnectome.org/
Glossary
- Cytoarchitectonics
-
The study of the spatial distribution pattern of neurons within the central nervous system; the size, shape, packing density and staining intensity of neuronal cell bodies in six layers are used to characterize a specific cytoarchitectural area.
- Clustering coefficient
-
The fraction of a node’s directly connected neighbours that are also neighbours of each other.
- Path length
-
The number of links connecting any two nodes in a network.
- Centrality
-
Measures that quantify the importance of a node or a link in a network.
- Modularity
-
A measure that quantifies the degree to which a network can be partitioned into subnetworks or modules.
- Small-world
-
A network property of high average clustering coefficient and short average shortest path length.
- Rich-club
-
A network has this property when nodes with high degree centrality are more densely interconnected between each other than expected.
- Global efficiency
-
The average inverse shortest path length in the network.
- Local efficiency
-
The inverse of the average shortest path length of all neighbours of the node and an alternative local connectivity metric to the clustering coefficient.
- Degree centrality
-
The number of links a node has to other nodes in a network.
- Epidemic spreading models
-
(ESM). Computational models simulating the spreading patterns of amyloid-β and tau from a preselected epicentre to different brain regions via structural connections.
- Euclidean distance matrix
-
A symmetric matrix, in which each element is computed by estimating the Euclidean distance between the centre coordinates of two brain regions.
- Network diffusion models
-
(NDMs). Computational models simulating Alzheimer disease progression on brain networks using a network heat equation.
Rights and permissions
About this article
Cite this article
Yu, M., Sporns, O. & Saykin, A.J. The human connectome in Alzheimer disease — relationship to biomarkers and genetics. Nat Rev Neurol 17, 545–563 (2021). https://doi.org/10.1038/s41582-021-00529-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41582-021-00529-1
This article is cited by
-
NMNAT2 supports vesicular glycolysis via NAD homeostasis to fuel fast axonal transport
Molecular Neurodegeneration (2024)
-
Odor identification score as an alternative method for early identification of amyloidogenesis in Alzheimer’s disease
Scientific Reports (2024)
-
The genetic architecture of multimodal human brain age
Nature Communications (2024)
-
Deep learning based computer aided diagnosis of Alzheimer’s disease: a snapshot of last 5 years, gaps, and future directions
Artificial Intelligence Review (2024)
-
A novel spatiotemporal graph convolutional network framework for functional connectivity biomarkers identification of Alzheimer’s disease
Alzheimer's Research & Therapy (2024)