Abstract
Biomolecular condensation arising through phase transitions has emerged as an essential organizational strategy that governs many aspects of cell biology. In particular, the role of phase transitions in the assembly of large, complex ribonucleoprotein (RNP) granules has become appreciated as an important regulator of RNA metabolism. In parallel, genetic, histopathological and cell and molecular studies have provided evidence that disturbance of phase transitions is an important driver of neurological diseases, notably amyotrophic lateral sclerosis (ALS), but most likely also other diseases. Indeed, our growing knowledge of the biophysics underlying biological phase transitions suggests that this process offers a unifying mechanism to explain the numerous and diverse disturbances in RNA metabolism that have been observed in ALS and some related diseases — specifically, that these diseases are driven by disturbances in the material properties of RNP granules. Here, we review the evidence for this hypothesis, emphasizing the reciprocal roles in which disease-related protein and disease-related RNA can lead to disturbances in the material properties of RNP granules and consequent pathogenesis. Additionally, we review evidence that implicates aberrant phase transitions as a contributing factor to a larger set of neurodegenerative diseases, including frontotemporal dementia, certain repeat expansion diseases and Alzheimer disease.
Key points
-
Intracellular phase transition describes a biophysical phenomenon in which macromolecules separate from their aqueous surroundings to form a functional compartment.
-
Biomolecular condensation arising through phase transitions is a cellular organizational strategy that governs many aspects of cell biology.
-
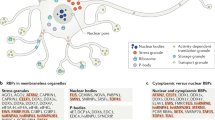
Among the largest biomolecular condensates are ribonucleoprotein (RNP) granules — membraneless organelles composed of RNA and protein — which regulate nearly all aspects of RNA metabolism.
-
Genetic, histopathological, cellular and molecular studies of amyotrophic lateral sclerosis (ALS) and related diseases have identified dysfunction in diverse aspects of RNA metabolism as a common theme.
-
Aberrant phase transitions and disturbances of the material properties of RNP granules offer a unifying mechanism for the numerous disturbances in RNA metabolism observed in ALS and related diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ghasemi, M. & Brown, R. H. Jr. Genetics of amyotrophic lateral sclerosis. Cold Spring Harb. Perspect. Med. 8, a024125 (2018).
Taylor, J. P., Brown, R. H. Jr & Cleveland, D. W. Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016).
Barmada, S. J. Linking RNA dysfunction and neurodegeneration in amyotrophic lateral sclerosis. Neurotherapeutics 12, 340–351 (2015).
Sreedharan, J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008).
Kabashi, E. et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574 (2008).
Vance, C. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009).
Kwiatkowski, T. J. Jr. et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009).
Kim, H. J. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013).
Liu, Q. et al. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology 87, 1763–1769 (2016).
Vieira, N. M. et al. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G). Hum. Mol. Genet. 23, 4103–4110 (2014).
Johnson, J. O. et al. Mutations in the matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17, 664–666 (2014).
Mackenzie, I. R. et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816 (2017).
Smith, B. N. et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci. Transl Med. 9, eaad9157 (2017).
Buchan, J. R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11, 1019–1030 (2014).
Gomes, E. & Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. https://doi.org/10.1074/jbc.TM118.001192 (2018).
Dundr, M. Nuclear bodies: multifunctional companions of the genome. Curr. Opin. Cell Biol. 24, 415–422 (2012).
Kiebler, M. A. & Bassell, G. J. Neuronal RNA granules: movers and makers. Neuron 51, 685–690 (2006).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Langdon, E. M. & Gladfelter, A. S. A new lens for RNA localization: liquid-liquid phase separation. Annu. Rev. Microbiol. 72, 255–271 (2018).
Zacharias, E. Über den Nucleolus. Botan. Zeit. 43, 257 (1885).
Walter, H. & Brooks, D. E. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 361, 135–139 (1995).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Altmeyer, M. et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 (2015).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Larson, A. G. et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017).
Cerase, A., Armaos, A., Cid, F., Avner, P. & Tartaglia, G. G. Xist lncRNA forms silencing granules that induce heterochromatin formation and repressive complexes recruitment by phase separation. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/351015v1 (2018).
Brangwynne, C. P., Mitchison, T. J. & Hyman, A. A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA 108, 4334–4339 (2011).
Sheu-Gruttadauria, J. & MacRae, I. J. Phase transitions in the assembly and function of human miRISC. Cell 173, 946–957 (2018).
Schmidt, H. B. & Gorlich, D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife 4, e04251 (2015).
Jiang, H. et al. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163, 108–122 (2015).
Banjade, S. & Rosen, M. K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 3, e04123 (2014).
Winton, M. J. et al. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 283, 13302–13309 (2008).
Johnson, B. S. et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 (2009).
Sun, Z. et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLOS Biol. 9, e1000614 (2011).
Kato, M. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012).
Van Treeck, B. & Parker, R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174, 791–802 (2018).
Leung, A. K. Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 205, 613–619 (2014).
McGurk, L. et al. Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717 (2018).
Patel, A. et al. ATP as a biological hydrotrope. Science 356, 753–756 (2017).
King, O. D., Gitler, A. D. & Shorter, J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462, 61–80 (2012).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Brangwynne, C. P., Tompa, P. & Pappu, R. V. Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899 (2015).
Wang, J. et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699 (2018).
Conicella, A. E., Zerze, G. H., Mittal, J. & Fawzi, N. L. ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 (2016).
Ryan, V. H. et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479 (2018).
Nelson, R. et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Hughes, M. P. et al. Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science 359, 698–701 (2018).
Guenther, E. L. et al. Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol. 25, 463–471 (2018).
Jain, S. et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016).
Protter, D. S. & Parker, R. Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016).
Kosik, K. S. & Krichevsky, A. M. The message and the messenger: delivering RNA in neurons. Sci. STKE 2002, e16 (2002).
Vogler, T. O. et al. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563, 508–513 (2018).
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006).
Salajegheh, M. et al. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve 40, 19–31 (2009).
Weihl, C. C. et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry 79, 1186–1189 (2008).
Irwin, D. J. et al. Frontotemporal lobar degeneration: defining phenotypic diversity through personalized medicine. Acta Neuropathol. 129, 469–491 (2015).
Kanekura, K. et al. Poly-dipeptides encoded by the C9orf72 repeats block global protein translation. Hum. Mol. Genet. 25, 1803–1813 (2016).
Deshaies, J. E. et al. TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis. Brain 141, 1320–1333 (2018).
Deng, H. X. et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann. Neurol. 67, 739–748 (2010).
Dormann, D. et al. ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import. EMBO J. 29, 2841–2857 (2010).
Lu, L. et al. Amyotrophic lateral sclerosis-linked mutant SOD1 sequesters Hu antigen R (HuR) and TIA-1-related protein (TIAR): implications for impaired post-transcriptional regulation of vascular endothelial growth factor. J. Biol. Chem. 284, 33989–33998 (2009).
Volkening, K., Leystra-Lantz, C., Yang, W., Jaffee, H. & Strong, M. J. TAR DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res. 1305, 168–182 (2009).
Liu-Yesucevitz, L. et al. TAR DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLOS ONE 5, e13250 (2010).
Elden, A. C. et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010).
Deng, H. X. et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 (2011).
Neumann, M. et al. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931 (2009).
Maruyama, H. et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226 (2010).
Schroder, R. et al. Mutant valosin-containing protein causes a novel type of frontotemporal dementia. Ann. Neurol. 57, 457–461 (2005).
Wilson, A. C., Dugger, B. N., Dickson, D. W. & Wang, D. S. TDP-43 in aging and Alzheimer’s disease — a review. Int. J. Clin. Exp. Pathol. 4, 147–155 (2011).
Schwab, C., Arai, T., Hasegawa, M., Yu, S. & McGeer, P. L. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J. Neuropathol. Exp. Neurol. 67, 1159–1165 (2008).
Uryu, K. et al. Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J. Neuropathol. Exp. Neurol. 67, 555–564 (2008).
Nakashima-Yasuda, H. et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 114, 221–229 (2007).
Lippa, C. F. et al. Transactive response DNA-binding protein 43 burden in familial Alzheimer disease and Down syndrome. Arch. Neurol. 66, 1483–1488 (2009).
Chanson, J. B. et al. TDP43-positive intraneuronal inclusions in a patient with motor neuron disease and Parkinson’s disease. Neurodegener. Dis. 7, 260–264 (2010).
Gallego-Iradi, M. C. et al. Subcellular localization of matrin 3 containing mutations associated with ALS and distal myopathy. PLOS ONE 10, e0142144 (2015).
Couthouis, J. et al. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl Acad. Sci. USA 108, 20881–20890 (2011).
Couthouis, J. et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 21, 2899–2911 (2012).
Freibaum, B. D., Chitta, R. K., High, A. A. & Taylor, J. P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 (2010).
Andersen, J. S. et al. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 (2002).
Shelkovnikova, T. A., Robinson, H. K., Troakes, C., Ninkina, N. & Buchman, V. L. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum. Mol. Genet. 23, 2298–2312 (2014).
Rajgor, D., Hanley, J. G. & Shanahan, C. M. Identification of novel nesprin-1 binding partners and cytoplasmic matrin-3 in processing bodies. Mol. Biol. Cell 27, 3894–3902 (2016).
Yamazaki, T. et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2, 799–806 (2012).
Wang, I. F., Reddy, N. M. & Shen, C. K. Higher order arrangement of the eukaryotic nuclear bodies. Proc. Natl Acad. Sci. USA 99, 13583–13588 (2002).
Ishihara, T. et al. Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum. Mol. Genet. 22, 4136–4147 (2013).
Murakami, T. et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690 (2015).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Guo, L. et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692 (2018).
Arnold, E. S. et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl Acad. Sci. USA 110, E736–E745 (2013).
Kawahara, Y. & Mieda-Sato, A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl Acad. Sci. USA 109, 3347–3352 (2012).
Coyne, A. N. et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet. 24, 6886–6898 (2015).
Alami, N. H. et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 81, 536–543 (2014).
Wang, I. F., Wu, L. S., Chang, H. Y. & Shen, C. K. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem. 105, 797–806 (2008).
Liao, Y.-C. et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Preprint at SSRN https://ssrn.com/abstract=3312723 (2019).
Kanai, Y., Dohmae, N. & Hirokawa, N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004).
Fujii, R. & Takumi, T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 118, 5755–5765 (2005).
Lopez-Erauskin, J. et al. ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron 100, 816–830 (2018).
Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 173, 720–734 (2018).
Buratti, E. & Baralle, F. E. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 (2001).
Hallier, M., Lerga, A., Barnache, S., Tavitian, A. & Moreau-Gachelin, F. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J. Biol. Chem. 273, 4838–4842 (1998).
Tollervey, J. R. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 (2011).
Polymenidou, M. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 (2011).
Lagier-Tourenne, C. et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15, 1488–1497 (2012).
Huelga, S. C. et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 1, 167–178 (2012).
Strong, M. J. et al. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell Neurosci. 35, 320–327 (2007).
Costessi, L., Porro, F., Iaconcig, A. & Muro, A. F. TDP-43 regulates beta-adducin (Add2) transcript stability. RNA Biol. 11, 1280–1290 (2014).
Colombrita, C. et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 287, 15635–15647 (2012).
Ling, J. P., Pletnikova, O., Troncoso, J. C. & Wong, P. C. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349, 650–655 (2015).
Schwartz, J. C. et al. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 26, 2690–2695 (2012).
Zhou, Y., Liu, S., Liu, G., Ozturk, A. & Hicks, G. G. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLOS Genet. 9, e1003895 (2013).
Guerrero, E. N. et al. TDP-43/FUS in motor neuron disease: complexity and challenges. Prog. Neurobiol. 145–146, 78–97 (2016).
Velazquez-Perez, L. C., Rodriguez-Labrada, R. & Fernandez-Ruiz, J. Spinocerebellar ataxia type 2: clinicogenetic aspects, mechanistic insights, and management approaches. Front. Neurol. 8, 472 (2017).
Becker, L. A. et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371 (2017).
Zhang, K. et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971 (2018).
Lee, K. H. et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788 (2016).
Bakthavachalu, B. et al. RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754–766 (2018).
Nonhoff, U. et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 18, 1385–1396 (2007).
Johnson, J. O. et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 (2010).
Fecto, F. et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 68, 1440–1446 (2011).
Wu, C. H. et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488, 499–503 (2012).
Nicolas, A. et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97, 1268–1283 (2018).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9orf72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9orf72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
Freischmidt, A. et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18, 631–636 (2015).
Bannwarth, S. et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 137, 2329–2345 (2014).
Smith, B. N. et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 84, 324–331 (2014).
Puls, I. et al. Mutant dynactin in motor neuron disease. Nat. Genet. 33, 455–456 (2003).
Greenway, M. J. et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 38, 411–413 (2006).
Chen, Y. Z. et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 74, 1128–1135 (2004).
Kaneb, H. M. et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum. Mol. Genet. 24, 1363–1373 (2015).
Buchan, J. R., Kolaitis, R. M., Taylor, J. P. & Parker, R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474 (2013).
Matus, S., Bosco, D. A. & Hetz, C. Autophagy meets fused in sarcoma-positive stress granules. Neurobiol. Aging 35, 2832–2835 (2014).
Dao, T. P. et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978 (2018).
Monahan, Z., Shewmaker, F. & Pandey, U. B. Stress granules at the intersection of autophagy and ALS. Brain Res. 1649, 189–200 (2016).
Thomas, M., Alegre-Abarrategui, J. & Wade-Martins, R. RNA dysfunction and aggrephagy at the centre of an amyotrophic lateral sclerosis/frontotemporal dementia disease continuum. Brain 136, 1345–1360 (2013).
Ash, P. E. et al. Unconventional translation of C9orf72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013).
Mann, D. M. et al. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9orf72. Acta Neuropathol. Commun. 1, 68 (2013).
Mori, K. et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 125, 413–423 (2013).
Mori, K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013).
Schludi, M. H. et al. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 130, 537–555 (2015).
Zu, T. et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA 108, 260–265 (2011).
Gendron, T. F. et al. Antisense transcripts of the expanded C9orf72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844 (2013).
Mackenzie, I. R. et al. Dipeptide repeat protein pathology in C9orf72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 126, 859–879 (2013).
Zu, T. et al. RAN proteins and RNA foci from antisense transcripts in C9orf72 ALS and frontotemporal dementia. Proc. Natl Acad. Sci. USA 110, E4968–E4977 (2013).
Todd, P. K. et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455 (2013).
Banez-Coronel, M. et al. RAN translation in Huntington disease. Neuron 88, 667–677 (2015).
Lin, Y. et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802 (2016).
Kwon, I. et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014).
Wen, X. et al. Antisense proline-arginine RAN dipeptides linked to C9orf72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225 (2014).
Freibaum, B. D. et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133 (2015).
Mizielinska, S. et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194 (2014).
Zhang, Y. J. et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142 (2018).
Prudencio, M. et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 18, 1175–1182 (2015).
Jovicic, A. et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 18, 1226–1229 (2015).
Zhang, K. et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015).
Burguete, A. S. et al. GGGGCC microsatellite RNA is neuritically localized, induces branching defects, and perturbs transport granule function. eLife 4, e08881 (2015).
Shi, K. Y. et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl Acad. Sci. USA 114, E1111–E1117 (2017).
Boeynaems, S. et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055 (2017).
Taneja, K. L., McCurrach, M., Schalling, M., Housman, D. & Singer, R. H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128, 995–1002 (1995).
Zhang, N. & Ashizawa, T. RNA toxicity and foci formation in microsatellite expansion diseases. Curr. Opin. Genet. Dev. 44, 17–29 (2017).
Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018).
Van Treeck, B. et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl Acad. Sci. USA 115, 2734–2739 (2018).
Khong, A. et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820 (2017).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
Lee, Y. B. et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 5, 1178–1186 (2013).
Wang, E. T. et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724 (2012).
Wojtkowiak-Szlachcic, A. et al. Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res. 43, 3318–3331 (2015).
Fratta, P. et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2, 1016 (2012).
Haeusler, A. R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 (2014).
Sareen, D. et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9orf72 repeat expansion. Sci. Transl Med. 5, 208ra149 (2013).
Donnelly, C. J. et al. RNA toxicity from the ALS/FTD C9orf72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013).
Xu, Z. et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl Acad. Sci. USA 110, 7778–7783 (2013).
Kim, H. J. & Taylor, J. P. Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron 96, 285–297 (2017).
Nousiainen, H. O. et al. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat. Genet. 40, 155–157 (2008).
Gasset-Rosa, F. et al. Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94, 48–57 (2017).
Hernandez-Vega, A. et al. Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 20, 2304–2312 (2017).
Wegmann, S. et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 (2018).
Zhang, X. et al. RNA stores tau reversibly in complex coacervates. PLOS Biol. 15, e2002183 (2017).
Ambadipudi, S., Biernat, J., Riedel, D., Mandelkow, E. & Zweckstetter, M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8, 275 (2017).
Ferreon, J. C. et al. Acetylation disfavors tau phase separation. Int. J. Mol. Sci. 19, E1360 (2018).
Kostylev, M. A. et al. Liquid and hydrogel phases of PrPC linked to conformation shifts and triggered by Alzheimer’s amyloid-beta oligomers. Mol. Cell 72, 426–443 (2018).
Anderson, P. & Kedersha, N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 (2008).
Decker, C. J. & Parker, R. P-Bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4, a012286 (2012).
Ramaswami, M., Taylor, J. P. & Parker, R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736 (2013).
Kedersha, N. et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 (2005).
Hoyle, N. P., Castelli, L. M., Campbell, S. G., Holmes, L. E. & Ashe, M. P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179, 65–74 (2007).
Parker, R. & Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 (2007).
Wilczynska, A., Aigueperse, C., Kress, M., Dautry, F. & Weil, D. The translational regulator CPEB1 provides a link between DCP1 bodies and stress granules. J. Cell Sci. 118, 981–992 (2005).
Mahboubi, H. & Stochaj, U. Nucleoli and stress granules: connecting distant relatives. Traffic 15, 1179–1193 (2014).
Trinkle-Mulcahy, L. & Sleeman, J. E. The Cajal body and the nucleolus: “in a relationship” or “it’s complicated”? RNA Biol. 14, 739–751 (2017).
Hofweber, M. et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719 (2018).
Wiltzius, J. J. et al. Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973–978 (2009).
Acknowledgements
J.P.T.’s research work is funded by the Howard Hughes Medical Institute and the St Jude Research Collaborative on the Biology of Membraneless Organelles.
Reviewer information
Nature Reviews Neurology thanks J. Rothstein and other anonymous reviewers for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Both authors contributed to researching data for the article, discussions of its content, writing and review or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
J.P.T. declares that he is a consultant for Third Rock Ventures. N.B.N. declares no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nedelsky, N.B., Taylor, J.P. Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat Rev Neurol 15, 272–286 (2019). https://doi.org/10.1038/s41582-019-0157-5
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41582-019-0157-5
This article is cited by
-
Glutathionylation on RNA-binding proteins: a regulator of liquid‒liquid phase separation in the pathogenesis of amyotrophic lateral sclerosis
Experimental & Molecular Medicine (2023)
-
Quantitative real-time in-cell imaging reveals heterogeneous clusters of proteins prior to condensation
Nature Communications (2023)
-
Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy
Journal of Human Genetics (2023)
-
Targeting the glycine-rich domain of TDP-43 with antibodies prevents its aggregation in vitro and reduces neurofilament levels in vivo
Acta Neuropathologica Communications (2023)
-
Learning the chemical grammar of biomolecular condensates
Nature Chemical Biology (2022)