Abstract
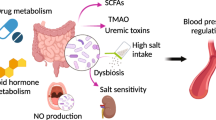
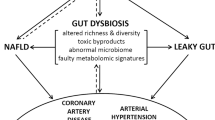
A large body of evidence has emerged in the past decade supporting a role for the gut microbiome in the regulation of blood pressure. The field has moved from association to causation in the last 5 years, with studies that have used germ-free animals, antibiotic treatments and direct supplementation with microbial metabolites. The gut microbiome can regulate blood pressure through several mechanisms, including through gut dysbiosis-induced changes in microbiome-associated gene pathways in the host. Microbiota-derived metabolites are either beneficial (for example, short-chain fatty acids and indole-3-lactic acid) or detrimental (for example, trimethylamine N-oxide), and can activate several downstream signalling pathways via G protein-coupled receptors or through direct immune cell activation. Moreover, dysbiosis-associated breakdown of the gut epithelial barrier can elicit systemic inflammation and disrupt intestinal mechanotransduction. These alterations activate mechanisms that are traditionally associated with blood pressure regulation, such as the renin–angiotensin–aldosterone system, the autonomic nervous system, and the immune system. Several methodological and technological challenges remain in gut microbiome research, and the solutions involve minimizing confounding factors, establishing causality and acting globally to improve sample diversity. New clinical trials, precision microbiome medicine and computational methods such as Mendelian randomization have the potential to enable leveraging of the microbiome for translational applications to lower blood pressure.
Key points
-
Clinical and experimental evidence show that changes in the gut microbiome are associated with and might lead to changes in blood pressure.
-
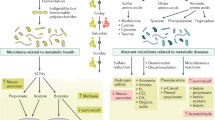
Metabolites produced by the gut microbiota are key mediators of the host–microbiota relationship that can drive changes to blood pressure; these metabolites can have immune-dependent and -independent effects.
-
Gut microbial-derived metabolites can be beneficial (for example, short-chain fatty acids, indole-3-lactic acid, arachidonic acid) or detrimental (for example, trimethylamine N-oxide) to blood pressure regulation.
-
Experimental hypertension is associated with disruption of the gut epithelial barrier and intestinal mechanotransduction; these might contribute to hypertension.
-
Gut microbiome modulation might represent a therapeutic approach to lowering blood pressure, for example, through the oral delivery of gut microbial metabolites such as short-chain fatty acids.
-
Addressing important challenges to gut microbiome research in the hypertension field, such as confounding factors, causality and global action to improve the diversity of samples, will accelerate discovery and translation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
GBD 2019 Risk Factors Collaborators. Global burden of 87 risk factors in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 396, 1223–1249 (2020).
Beaney, T. et al. May measurement month 2019: the global blood pressure screening campaign of the International Society of Hypertension. Hypertension 76, 333–341 (2020).
GBD 2019 Risk Factors Collaborators. Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: a pooled analysis of 1201 population-representative studies with 104 million participants. Lancet 398, 957–980 (2021).
Oparil, S. et al. Hypertension. Nat. Rev. Dis. Prim. 4, 18014 (2018).
Page, I. H. Pathogenesis of arterial hypertension. J. Am. Med. Assoc. 140, 451–458 (1949).
Kolifarhood, G. et al. Heritability of blood pressure traits in diverse populations: a systematic review and meta-analysis. J. Hum. Hypertens. 33, 775–785 (2019).
Evangelou, E. et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425 (2018).
Marques, F. Z. Missing heritability of hypertension and our microbiome. Circulation 138, 1381–1383 (2018).
Marques, F. Z., Mackay, C. R. & Kaye, D. M. Beyond gut feelings: how the gut microbiota regulates blood pressure. Nat. Rev. Cardiol. 15, 20–32 (2018).
Harrison, D. G., Coffman, T. M. & Wilcox, C. S. Pathophysiology of hypertension: the mosaic theory and beyond. Circ. Res. 128, 847–863 (2021). This comprehensive review introduced a revised version of the mosaic theory of hypertension that acknowledges the role of the gut microbiome in the pathophysiology of hypertension.
Rogler, G. & Rosano, G. The heart and the gut. Eur. Heart J. 35, 426–430 (2014).
Beale, A. L. et al. The gut microbiome of heart failure with preserved ejection fraction. J. Am. Heart Assoc. 10, e020654 (2021).
Wang, B. et al. Blood pressure-lowering effects of GLP-1 receptor agonists exenatide and liraglutide: a meta-analysis of clinical trials. Diabetes Obes. Metab. 15, 737–749 (2013).
Richards, E. M., Li, J., Stevens, B. R., Pepine, C. J. & Raizada, M. K. Gut microbiome and neuroinflammation in hypertension. Circ. Res. 130, 401–417 (2022).
Pugh, D., Gallacher, P. J. & Dhaun, N. Management of hypertension in chronic kidney disease. Drugs 79, 365–379 (2019).
Yang, T., Richards, E. M., Pepine, C. J. & Raizada, M. K. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat. Rev. Nephrol. 14, 442–456 (2018).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788 (2008).
Ramakrishna, B. S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 28 (Suppl. 4), 9–17 (2013).
Ducarmon, Q. R. et al. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol. Mol. Biol. Rev. https://doi.org/10.1128/MMBR.00007-19 (2019).
Sanidad, K. Z. & Zeng, M. Y. Neonatal gut microbiome and immunity. Curr. Opin. Microbiol. 56, 30–37 (2020).
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009).
Berg, G. et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 103 (2020).
Sanna, S., Kurilshikov, A., van der Graaf, A., Fu, J. & Zhernakova, A. Challenges and future directions for studying effects of host genetics on the gut microbiome. Nat. Genet. 54, 100–106 (2022). This Perspective discusses challenges and future directions for genetic analysis of the microbiome.
Koponen, K. K. et al. Associations of healthy food choices with gut microbiota profiles. Am. J. Clin. Nutr. 114, 605–616 (2021).
Leeming, E. R., Johnson, A. J., Spector, T. D. & Le Roy, C. I. Effect of diet on the gut microbiota: rethinking intervention duration. Nutrients https://doi.org/10.3390/nu11122862 (2019).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). This study provided the first evidence that shifting dietary nutrients can rapidly, broadly and consistently alter gut microbiome composition in humans.
Reynolds, A. et al. Carbohydrate quality and human health: a series of systematic reviews and meta-analyses. Lancet 393, 434–445 (2019).
Xu, C. & Marques, F. Z. How dietary fibre, acting via the gut microbiome, lowers blood pressure. Curr. Hypertens. Rep. 24, 509–521 (2022).
De Filippis, F. et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65, 1812–1821 (2016).
Gill, P. A., van Zelm, M. C., Muir, J. G. & Gibson, P. R. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment. Pharmacol. Ther. 48, 15–34 (2018).
Korpela, K. Diet, microbiota, and metabolic health: trade-off between saccharolytic and proteolytic fermentation. Annu. Rev. Food Sci. Technol. 9, 65–84 (2018).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
So, D., Gibson, P. R., Muir, J. G. & Yao, C. K. Dietary fibres and IBS: translating functional characteristics to clinical value in the era of personalised medicine. Gut 70, 2383–2394 (2021).
Goodrich, J. K. et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743 (2016).
Bonder, M. J. et al. The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412 (2016).
Grieneisen, L. et al. Gut microbiome heritability is nearly universal but environmentally contingent. Science 373, 181–186 (2021).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Hall, A. B., Tolonen, A. C. & Xavier, R. J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 18, 690–699 (2017).
Qin, Y. et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 54, 134–142 (2022). This large-scale study demonstrated the combined effect of host genetics and diet on the human gut microbiota.
Lopera-Maya, E. A. et al. Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch Microbiome Project. Nat. Genet. 54, 143–151 (2022).
Hu, X., Li, J., Fu, M., Zhao, X. & Wang, W. The JAK/STAT signaling pathway: from bench to clinic. Signal. Transduct. Target. Ther. 6, 402 (2021).
Groot, H. E. et al. Genetically determined ABO blood group and its associations with health and disease. Arterioscler. Thromb. Vasc. Biol. 40, 830–838 (2020).
Vijay, A. & Valdes, A. M. Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr. 76, 489–501 (2022).
Nakai, M. et al. Essential hypertension is associated with changes in gut microbial metabolic pathways: a multisite analysis of ambulatory blood pressure. Hypertension 78, 804–815 (2021).
Kim, S. et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin. Sci. 132, 701–718 (2018).
Li, J. et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5, 14 (2017). The first faecal microbiome transplantation study to show the causal role of the gut microbiome in blood pressure regulation.
Yang, T. et al. Gut dysbiosis is linked to hypertension. Hypertension 65, 1331–1340 (2015). The first study to describe gut dysbiosis in experimental and human hypertension.
Palmu, J. et al. Association between the gut microbiota and blood pressure in a population cohort of 6953 individuals. J. Am. Heart Assoc. 9, e016641 (2020).
Huart, J. et al. Gut microbiota and fecal levels of short-chain fatty acids differ upon 24-hour blood pressure levels in men. Hypertension https://doi.org/10.1161/HYPERTENSIONAHA.118.12588 (2019).
Muralitharan, R. R. et al. Microbial peer pressure: the role of the gut microbiota in hypertension and its complications. Hypertension 76, 1674–1687 (2020).
Qv, L. et al. Methods for establishment and maintenance of germ-free rat models. Front. Microbiol. 11, 1148 (2020).
Chen, X. et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut 69, 513–522 (2020).
Yang, T. et al. Sustained captopril-induced reduction in blood pressure is associated with alterations in gut-brain axis in the spontaneously hypertensive rat. J. Am. Heart Assoc. 8, e010721 (2019).
Robles-Vera, I. et al. Changes to the gut microbiota induced by losartan contributes to its antihypertensive effects. Br. J. Pharmacol. 177, 2006–2023 (2020).
Yang, T. et al. Identification of a gut commensal that compromises the blood pressure-lowering effect of ester angiotensin-converting enzyme inhibitors. Hypertension https://doi.org/10.1161/HYPERTENSIONAHA.121.18711 (2022). This study showed that the gut microbiome can affect the efficacy of anti-hypertensive medications.
Nicholson, J. K. et al. Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012).
Marques, F. Z. et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 135, 964–977 (2017). This study provided the first evidence that prebiotic fibre has a blood pressure-lowering effect by modulating the gut microbiota and the short-chain fatty acid acetate.
Tang, T. W. H. et al. Loss of gut microbiota alters immune system composition and cripples postinfarction cardiac repair. Circulation 139, 647–659 (2019).
Stavropoulou, E. et al. Focus on the gut-kidney axis in health and disease. Front. Med. 7, 620102 (2020).
Morais, L. H., Schreiber, H. L. T. & Mazmanian, S. K. The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 19, 241–255 (2021).
Peh, A., O’Donnell, J. A., Broughton, B. R. S. & Marques, F. Z. Gut microbiota and their metabolites in stroke: a double-edged sword. Stroke 53, 1788–1801 (2022).
Gebrayel, P. et al. Microbiota medicine: towards clinical revolution. J. Transl. Med. 20, 111 (2022).
Drummond, G. R., Vinh, A., Guzik, T. J. & Sobey, C. G. Immune mechanisms of hypertension. Nat. Rev. Immunol. 19, 517–532 (2019).
Karbach, S. H. et al. Gut microbiota promote angiotensin II-induced arterial hypertension and vascular dysfunction. J. Am. Heart Assoc. https://doi.org/10.1161/JAHA.116.003698 (2016).
Avery, E. G. et al. Quantifying the impact of gut microbiota on inflammation and hypertensive organ damage. Cardiovasc. Res. https://doi.org/10.1093/cvr/cvac121 (2022).
Joe, B. et al. Microbiota introduced to germ-free rats restores vascular contractility and blood pressure. Hypertension 76, 1847–1855 (2020).
Mortensen, F. V., Nielsen, H., Mulvany, M. J. & Hessov, I. Short chain fatty acids dilate isolated human colonic resistance arteries. Gut 31, 1391–1394 (1990).
Kaye, D. M. et al. Deficiency of prebiotic fiber and insufficient signaling through gut metabolite-sensing receptors leads to cardiovascular disease. Circulation 141, 1393–1403 (2020). This study linked the lack of prebiotic dietary fibre to the development of hypertension and demonstrated the cardioprotective role of short-chain fatty acid receptors.
Jama, H. A. et al. Prebiotic intervention with HAMSAB in untreated essential hypertensive patients assessed in a phase II randomized trial. Nat. Cardiovasc. Res. https://doi.org/10.1038/s44161-022-00197-4 (2023).
Wang, L. et al. Sodium butyrate suppresses angiotensin II-induced hypertension by inhibition of renal (pro)renin receptor and intrarenal renin-angiotensin system. J. Hypertens. 35, 1899–1908 (2017).
Onyszkiewicz, M. et al. Butyric acid, a gut bacteria metabolite, lowers arterial blood pressure via colon-vagus nerve signaling and GPR41/43 receptors. Pflugers Arch. 471, 1441–1453 (2019).
Bartolomaeus, H. et al. The short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation 139, 1407–1421 (2019). This study highlighted the role of T regulatory cell homeostasis, driven by propionate, in the reduction of inflammation and fibrosis.
Guzik, T. J. et al. Role of the T cell in the genesis of angiotensin II induced hypertension and vascular dysfunction. J. Exp. Med. 204, 2449–2460 (2007).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Barhoumi, T. et al. T regulatory lymphocytes prevent angiotensin II-induced hypertension and vascular injury. Hypertension 57, 469–476 (2011).
Macia, L. et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6, 6734 (2015).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009).
Natarajan, N. et al. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G-protein coupled receptor 41. Physiol. Genomics 48, 826–834 (2016).
Rhys-Jones, D. et al. Microbial interventions to control and reduce blood pressure in Australia (MICRoBIA): rationale and design of a double-blinded randomised cross-over placebo controlled trial. Trials 22, 496 (2021).
Hu, J. et al. Enteric dysbiosis-linked gut barrier disruption triggers early renal injury induced by chronic high salt feeding in mice. Exp. Mol. Med. 49, e370 (2017).
Mell, B. et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol. Genomics 47, 187–197 (2015).
Ferguson, J. F. et al. High dietary salt-induced dendritic cell activation underlies microbial dysbiosis-associated hypertension. JCI Insight https://doi.org/10.1172/jci.insight.126241 (2019).
Kirabo, A. et al. DC isoketal-modified proteins activate T cells and promote hypertension. J. Clin. Invest. 124, 4642–4656 (2014). This study showed that salt drives T cell activation via isoketal-modified proteins on dendritic cells to increase blood pressure.
Chen, L. et al. Modest sodium reduction increases circulating short-chain fatty acids in untreated hypertensives: a randomized, double-blind, placebo-controlled trial. Hypertension 76, 73–79 (2020).
Wilck, N. et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551, 585–589 (2017).
Toral, M. et al. The probiotic Lactobacillus coryniformis CECT5711 reduces the vascular pro-oxidant and pro-inflammatory status in obese mice. Clin. Sci. 127, 33–45 (2014).
Yan, X. et al. Intestinal flora modulates blood pressure by regulating the synthesis of intestinal-derived corticosterone in high salt-induced hypertension. Circ. Res. https://doi.org/10.1161/CIRCRESAHA.119.316394 (2020).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Witkowski, M., Weeks, T. L. & Hazen, S. L. Gut microbiota and cardiovascular disease. Circ. Res. 127, 553–570 (2020).
Jiang, S. et al. Gut microbiota dependent trimethylamine N-oxide aggravates angiotensin II-induced hypertension. Redox Biol. 46, 102115 (2021).
Brunt, V. E. et al. Gut microbiome-derived metabolite trimethylamine N-oxide induces aortic stiffening and increases systolic blood pressure with aging in mice and humans. Hypertension 78, 499–511 (2021).
Roberts, A. B. et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 24, 1407–1417 (2018).
Yao, L. et al. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife https://doi.org/10.7554/eLife.37182 (2018).
Mistry, R. H., Verkade, H. J. & Tietge, U. J. Reverse cholesterol transport is increased in germ-free mice-brief report. Arterioscler. Thromb. Vasc. Biol. 37, 419–422 (2017).
Jones, B. V., Begley, M., Hill, C., Gahan, C. G. & Marchesi, J. R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl Acad. Sci. USA 105, 13580–13585 (2008).
Zhu, Q. et al. Moderation of gut microbiota and bile acid metabolism by chlorogenic acid improves high-fructose-induced salt-sensitive hypertension in mice. Food Funct. 13, 6987–6999 (2022).
Ji, C. G. et al. Bile acid receptor TGR5 overexpression is associated with decreased intestinal mucosal injury and epithelial cell proliferation in obstructive jaundice. Transl. Res. 182, 88–102 (2017).
Makishima, M. et al. Identification of a nuclear receptor for bile acids. Science 284, 1362–1365 (1999).
Shi, H. et al. Restructuring the gut microbiota by intermittent fasting lowers blood pressure. Circ. Res. 128, 1240–1254 (2021).
Li, C., Li, J., Weng, X., Lan, X. & Chi, X. Farnesoid X receptor agonist CDCA reduces blood pressure and regulates vascular tone in spontaneously hypertensive rats. J. Am. Soc. Hypertens. 9, 507–516 e507 (2015).
Campbell, C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020).
Santisteban, M. M. et al. Hypertension-linked pathophysiological alterations in the gut. Circ. Res. 120, 312–323 (2017). This work provided the first evidence of gut epithelial disruption in experimental hypertension.
Toral, M. et al. Critical role of the interaction gut microbiota - sympathetic nervous system in the regulation of blood pressure. Front. Physiol. 10, 231 (2019).
Helander, H. F. & Fandriks, L. Surface area of the digestive tract — revisited. Scand. J. Gastroenterol. 49, 681–689 (2014).
Odenwald, M. A. & Turner, J. R. The intestinal epithelial barrier: a therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 14, 9–21 (2017). This comprehensive Review discussed the role of intestinal epithelial barrier function in the pathogenesis of intestinal and systemic diseases.
Ghosh, S., Whitley, C. S., Haribabu, B. & Jala, V. R. Regulation of intestinal barrier function by microbial metabolites. Cell Mol. Gastroenterol. Hepatol. 11, 1463–1482 (2021).
Ghosh, S. S., Wang, J., Yannie, P. J. & Ghosh, S. Intestinal barrier dysfunction, LPS translocation, and disease development. J. Endocr. Soc. 4, bvz039 (2020).
Hansson, G. C. Mucins and the microbiome. Annu. Rev. Biochem. 89, 769–793 (2020).
Niessen, C. M. Tight junctions/adherens junctions: basic structure and function. J. Invest. Dermatol. 127, 2525–2532 (2007).
Furuse, M. et al. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 123, 1777–1788 (1993).
Van Itallie, C. M. & Anderson, J. M. Claudins and epithelial paracellular transport. Annu. Rev. Physiol. 68, 403–429 (2006).
Hartmann, C., Schwietzer, Y. A., Otani, T., Furuse, M. & Ebnet, K. Physiological functions of junctional adhesion molecules (JAMs) in tight junctions. Biochim. Biophys. Acta Biomembr. 1862, 183299 (2020).
Odenwald, M. A. et al. The scaffolding protein ZO-1 coordinates actomyosin and epithelial apical specializations in vitro and in vivo. J. Biol. Chem. 293, 17317–17335 (2018).
Hartsock, A. & Nelson, W. J. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta 1778, 660–669 (2008).
Zuo, L., Kuo, W. T. & Turner, J. R. Tight junctions as targets and effectors of mucosal immune homeostasis. Cell Mol. Gastroenterol. Hepatol. 10, 327–340 (2020).
Pabst, O. & Slack, E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. 13, 12–21 (2020).
Mabbott, N. A., Donaldson, D. S., Ohno, H., Williams, I. R. & Mahajan, A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 6, 666–677 (2013).
Schneider, C., O’Leary, C. E. & Locksley, R. M. Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 19, 584–593 (2019).
Zheng, D., Liwinski, T. & Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506 (2020).
Bischoff, S. C. et al. Intestinal permeability — a new target for disease prevention and therapy. BMC Gastroenterol. 14, 189 (2014).
Chelakkot, C., Ghim, J. & Ryu, S. H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 50, 1–9 (2018).
Sharma, R. K. et al. Microglial cells impact gut microbiota and gut pathology in angiotensin II-induced hypertension. Circ. Res. 124, 727–736 (2019).
Ntlahla, E. E., Mfengu, M. M., Engwa, G. A., Nkeh-Chungag, B. N. & Sewani-Rusike, C. R. Gut permeability is associated with hypertension and measures of obesity but not with endothelial dysfunction in South African youth. Afr. Health Sci. 21, 1172–1184 (2021).
Li, C. et al. Risk factors for intestinal barrier impairment in patients with essential hypertension. Front. Med. 7, 543698 (2020).
Pearce, K., Estanislao, D., Fareed, S. & Tremellen, K. Metabolic endotoxemia, feeding studies and the use of the limulus amebocyte (LAL) assay; is it fit for purpose? Diagnostics https://doi.org/10.3390/diagnostics10060428 (2020).
Mercado-Perez, A. & Beyder, A. Gut feelings: mechanosensing in the gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 19, 283–296 (2022). This Review described the role of gastrointestinal mechanosensation in gut function.
Farrugia, G. et al. A mechanosensitive calcium channel in human intestinal smooth muscle cells. Gastroenterology 117, 900–905 (1999).
Romero, S., Le Clainche, C. & Gautreau, A. M. Actin polymerization downstream of integrins: signaling pathways and mechanotransduction. Biochem. J. 477, 1–21 (2020).
Schwayer, C. et al. Mechanosensation of tight junctions depends on ZO-1 phase separation and flow. Cell 179, 937–952 e918 (2019).
Joshi, V., Strege, P. R., Farrugia, G. & Beyder, A. Mechanotransduction in gastrointestinal smooth muscle cells: role of mechanosensitive ion channels. Am. J. Physiol. Gastrointest. Liver Physiol. 320, G897–G906 (2021).
Hahn, C. & Schwartz, M. A. Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 10, 53–62 (2009).
Ye, G. J., Nesmith, A. P. & Parker, K. K. The role of mechanotransduction on vascular smooth muscle myocytes’ [corrected] cytoskeleton and contractile function. Anat. Rec. 297, 1758–1769 (2014).
Zeng, W. Z. et al. PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science 362, 464–467 (2018).
Williams, E. K. et al. Sensory neurons that detect stretch and nutrients in the digestive system. Cell 166, 209–221 (2016).
Won, K. J., Sanders, K. M. & Ward, S. M. Interstitial cells of Cajal mediate mechanosensitive responses in the stomach. Proc. Natl Acad. Sci. USA 102, 14913–14918 (2005).
Muller, P. A. et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 158, 300–313 (2014).
Page, A. J. & Li, H. Meal-sensing signaling pathways in functional dyspepsia. Front. Syst. Neurosci. 12, 10 (2018).
Neshatian, L. et al. Ranolazine inhibits voltage-gated mechanosensitive sodium channels in human colon circular smooth muscle cells. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G506–G512 (2015).
Strege, P. R. et al. Irritable bowel syndrome patients have SCN5A channelopathies that lead to decreased NaV1.5 current and mechanosensitivity. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G494–G503 (2018).
Tustumi, F., Morrell, A. L. G., Szor, D. J. & Dias, A. R. Achalasia: a mechanical and sensitivity disorder. United European Gastroenterol. J. 8, 1126–1127 (2020).
Tomsen, N. et al. Acute and subacute effects of oropharyngeal sensory stimulation with TRPV1 agonists in older patients with oropharyngeal dysphagia: a biomechanical and neurophysiological randomized pilot study. Ther. Adv. Gastroenterol. 12, 1756284819842043 (2019).
Stewart, D. C. et al. Hypertension-linked mechanical changes of rat gut. Acta Biomater. 45, 296–302 (2016).
Vandeputte, D. et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65, 57–62 (2016).
Wolter, M. et al. Leveraging diet to engineer the gut microbiome. Nat. Rev. Gastroenterol. Hepatol. 18, 885–902 (2021).
Wortelboer, K., Nieuwdorp, M. & Herrema, H. Fecal microbiota transplantation beyond Clostridioides difficile infections. EBioMedicine 44, 716–729 (2019).
Fan, L. et al. Effect of fecal microbiota transplantation on primary hypertension and the underlying mechanism of gut microbiome restoration: protocol of a randomized, blinded, placebo-controlled study. Trials 23, 178 (2022).
Ke, S., Weiss, S. T. & Liu, Y. Y. Rejuvenating the human gut microbiome. Trends Mol. Med. https://doi.org/10.1016/j.molmed.2022.05.005 (2022).
Zhang, T. et al. Washed microbiota transplantation vs. manual fecal microbiota transplantation: clinical findings, animal studies and in vitro screening. Protein Cell 11, 251–266 (2020).
Zhong, H. J. et al. Washed microbiota transplantation lowers blood pressure in patients with hypertension. Front. Cell Infect. Microbiol. 11, 679624 (2021).
Bishehsari, F., Voigt, R. M. & Keshavarzian, A. Circadian rhythms and the gut microbiota: from the metabolic syndrome to cancer. Nat. Rev. Endocrinol. 16, 731–739 (2020).
Mirzayi, C. et al. Reporting guidelines for human microbiome research: the STORMS checklist. Nat. Med. 27, 1885–1892 (2021).
Marques, F. Z. et al. Guidelines for transparency on gut microbiome studies in essential and experimental hypertension. Hypertension 74, 1279–1293 (2019). A complete guideline for conducting gut microbiome studies in experimental and clinical hypertension.
Dinakis, E. et al. Association between the gut microbiome and their metabolites with human blood pressure variability. Hypertension 79, 1690–1701 (2022).
Zhang, X., Li, L., Butcher, J., Stintzi, A. & Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 7, 154 (2019).
Kurilshikov, A. et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165 (2021).
Jackson, M. A. et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat. Commun. 9, 2655 (2018).
Nearing, J. T., Comeau, A. M. & Langille, M. G. I. Identifying biases and their potential solutions in human microbiome studies. Microbiome 9, 113 (2021).
Bell, G., Hey, T. & Szalay, A. Computer science. Beyond the data deluge. Science 323, 1297–1298 (2009).
Parks, D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794 (2022).
Durazzi, F. et al. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 11, 3030 (2021).
Brussow, H. Problems with the concept of gut microbiota dysbiosis. Microb. Biotechnol. 13, 423–434 (2020).
Wei, S., Bahl, M. I., Baunwall, S. M. D., Hvas, C. L. & Licht, T. R. Determining gut microbial dysbiosis: a review of applied indexes for assessment of intestinal microbiota imbalances. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.00395-21 (2021).
Nearing, J. T. et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 13, 342 (2022).
Hughes, D. A. et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat. Microbiol. 5, 1079–1087 (2020).
Wade, K. H. & Hall, L. J. Improving causality in microbiome research: can human genetic epidemiology help? Wellcome Open Res. 4, 199 (2019).
Forster, S. C. et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat. Biotechnol. 37, 186–192 (2019).
Abdill, R. J., Adamowicz, E. M. & Blekhman, R. Public human microbiome data are dominated by highly developed countries. PLoS Biol. 20, e3001536 (2022). This study investigated the global pattern of microbiome-associated diseases and demonstrated an imbalanced global representation of participants in microbiome studies.
Walejko, J. M. et al. Gut microbiota and serum metabolite differences in African Americans and White Americans with high blood pressure. Int. J. Cardiol. 271, 336–339 (2018).
Sun, S. et al. Gut microbiota composition and blood pressure. Hypertension 73, 998–1006 (2019).
Popejoy, A. B. & Fullerton, S. M. Genomics is failing on diversity. Nature 538, 161–164 (2016).
Gupta, V. K., Paul, S. & Dutta, C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 8, 1162 (2017).
Mei, X. et al. Beyond the gastrointestinal tract: oral and sex-specific skin microbiota are associated with hypertension in rats with genetic disparities. Physiol. Genomics 54, 242–250 (2022).
Beale, A. L., Kaye, D. M. & Marques, F. Z. The role of the gut microbiome in sex differences in arterial pressure. Biol. Sex. Differ. 10, 22 (2019).
Konop, M. et al. Enalapril decreases rat plasma concentration of TMAO, a gut bacteria-derived cardiovascular marker. Biomarkers 23, 380–385 (2018).
Yoo, H. H., Kim, I. S., Yoo, D. H. & Kim, D. H. Effects of orally administered antibiotics on the bioavailability of amlodipine: gut microbiota-mediated drug interaction. J. Hypertens. 34, 156–162 (2016).
Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R. & Goodman, A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019).
Brocker, C. N. et al. Metabolomic profiling of metoprolol hypertension treatment reveals altered gut microbiota-derived urinary metabolites. Hum. Genomics 14, 10 (2020).
Trott, D. W. et al. Oligoclonal CD8+ T cells play a critical role in the development of hypertension. Hypertension 64, 1108–1115 (2014).
Madhur, M. S. et al. Interleukin 17 promotes angiotensin II-induced hypertension and vascular dysfunction. Hypertension 55, 500–507 (2010).
Norlander, A. E. et al. Interleukin-17A regulates renal sodium transporters and renal injury in angiotensin II-induced hypertension. Hypertension 68, 167–174 (2016).
Vinh, A. et al. Inhibition and genetic ablation of the B7/CD28 T-cell costimulation axis prevents experimental hypertension. Circulation 122, 2529–2537 (2010).
Barbaro, N. R. et al. Dendritic cell amiloride-sensitive channels mediate sodium-induced inflammation and hypertension. Cell Rep. 21, 1009–1020 (2017).
Van Beusecum, J. P. et al. High salt activates CD11c+ antigen-presenting cells via SGK (Serum Glucocorticoid Kinase) 1 to promote renal inflammation and salt-sensitive hypertension. Hypertension 74, 555–563 (2019).
Ko, E. A. et al. Resistance artery remodeling in deoxycorticosterone acetate-salt hypertension is dependent on vascular inflammation: evidence from m-CSF-deficient mice. Am. J. Physiol. Heart Circ. Physiol. 292, H1789–H1795 (2007).
Pluznick, J. L. et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl Acad. Sci. USA 110, 4410–4415 (2013).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Brown, A. J. et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278, 11312–11319 (2003).
Tazoe, H. et al. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed. Res. 30, 149–156 (2009).
Karaki, S. et al. Expression of the short-chain fatty acid receptor, GPR43, in the human colon. J. Mol. Histol. 39, 135–142 (2008).
Acknowledgements
J.A.O. is supported by a National Health and Medical Research Council Early Career Fellowship (1124288). F.Z.M. is supported by a Senior Medical Research Fellowship from the Sylvia and Charles Viertel Charitable Foundation Fellowship and a National Heart Foundation Future Leader Fellowship (105663). G.M. is partially supported by NHMRC grant GNT2013468.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, made substantial contributions to discussions of the content and wrote, reviewed or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Nephrology thanks J. Cai, I. Robles-Vera and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Genome Taxonomy Database: https://gtdb.ecogenomic.org/
Glossary
- Colonization resistance
-
The ability of the commensal microbiota to limit the expansion of pathogens and exogenous microorganisms.
- Coprophagy
-
The ingestion of faeces; in this context by rodents.
- Interstitial cells of Cajal
-
Mesenchymal cells located in the intestine that mediate contractility.
- Microfold (M) cells
-
Specialized intestinal cells, located in the Peyer’s patches and other mucosa-associated lymphoid tissues, which sample pathogens or antigens from the intestinal lumen to the sub-epithelium.
- Tuft cells
-
Chemosensory cells located in the gut epithelial layer.
- Zero-inflation
-
Microbiome data typically contain several taxa (metagenome-assembled genomes or operational taxonomic units) that are only present in some samples, resulting in many taxa with zero counts.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
O’Donnell, J.A., Zheng, T., Meric, G. et al. The gut microbiome and hypertension. Nat Rev Nephrol 19, 153–167 (2023). https://doi.org/10.1038/s41581-022-00654-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41581-022-00654-0
This article is cited by
-
Microbiota-derived acetate attenuates neuroinflammation in rostral ventrolateral medulla of spontaneously hypertensive rats
Journal of Neuroinflammation (2024)
-
Influence of gut microbiome on metabolic diseases: a new perspective based on microgravity
Journal of Diabetes & Metabolic Disorders (2024)
-
Pathophysiology of primary hypertension in children and adolescents
Pediatric Nephrology (2024)
-
Gut microbiome profile of Chinese hypertension patients with and without type 2 diabetes mellitus
BMC Microbiology (2023)
-
Impact of gut microbiome on the renin-aldosterone system: Shika-machi Super Preventive Health Examination results
Hypertension Research (2023)