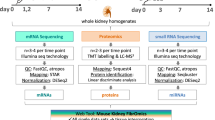
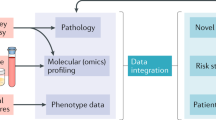
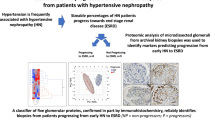
Abstract
Kidney research is entering an era of ‘big data’ and molecular omics data can provide comprehensive insights into the molecular footprints of cells. In contrast to transcriptomics, proteomics and metabolomics generate data that relate more directly to the pathological symptoms and clinical parameters observed in patients. Owing to its complexity, the proteome still holds many secrets, but has great potential for the identification of drug targets. Proteomics can provide information about protein synthesis, modification and degradation, as well as insight into the physical interactions between proteins, and between proteins and other biomolecules. Thus far, proteomics in nephrology has largely focused on the discovery and validation of biomarkers, but the systematic analysis of the nephroproteome can offer substantial additional insights, including the discovery of mechanisms that trigger and propagate kidney disease. Moreover, proteome acquisition might provide a diagnostic tool that complements the assessment of a kidney biopsy sample by a pathologist. Such applications are becoming increasingly feasible with the development of high-throughput and high-coverage technologies, such as versatile mass spectrometry-based techniques and protein arrays, and encourage further proteomics research in nephrology.
Key points
-
Proteomics analyses all the physical and chemical properties of proteins using a wide variety of technical approaches.
-
The versatility of proteome capture modes gives deep mechanistic insights into clinical samples and animal models of glomerular disease.
-
Both biomarker discovery and biopsy interrogation are key applications that have been propelled by increasingly simplified and sensitive sample preparation methods.
-
Given their diversified landscape, the analysis of post-translational modifications can uncover novel mechanisms of protein regulation.
-
Computational integration and mathematical modelling of molecular processes — tools largely developed for oncology — can be adapted to kidney proteomics research.
-
An increasingly clinical focus of proteomics analyses in combination with other omics technologies will aid or even drive novel discoveries of disease mechanisms and biomarkers in kidney disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lindenmeyer, M. T. & Kretzler, M. Renal biopsy-driven molecular target identification in glomerular disease. Pflugers Arch. 469, 1021–1028 (2017).
Kiryluk, K. et al. Precision medicine for acute kidney injury (AKI): redefining AKI by agnostic kidney tissue interrogation and genetics. Semin. Nephrol. 38, 40–51 (2018).
Saez-Rodriguez, J., Rinschen, M. M., Floege, J. & Kramann, R. Big science and big data in nephrology. Kidney Int. 95, 1326–1337 (2019).
Smith, L. M. & Kelleher, N. L. Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 (2013).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Olsen, J. V. & Mann, M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell Proteom. 12, 3444–3452 (2013).
Overington, J. P., Al-Lazikani, B. & Hopkins, A. L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Bache, N. et al. A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics. Mol. Cell Proteom. 17, 2284–2296 (2018).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Wilhelm, M. et al. Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 (2014).
Kim, M.-S. et al. A draft map of the human proteome. Nature 509, 575–581 (2014).
Hoyer, K. J. R., Dittrich, S., Bartram, M. P. & Rinschen, M. M. Quantification of molecular heterogeneity in kidney tissue by targeted proteomics. J. Proteom. 193, 85–92 (2019).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Hayek, S. S. et al. A tripartite complex of suPAR, APOL1 risk variants and αvβ3 integrin on podocytes mediates chronic kidney disease. Nat. Med. 23, 945–953 (2017).
Beck, L. H. et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N. Engl. J. Med. 361, 11–21 (2009).
Hobeika, L., Barati, M. T., Caster, D. J., McLeish, K. R. & Merchant, M. L. Characterization of glomerular extracellular matrix by proteomic analysis of laser-captured microdissected glomeruli. Kidney Int. 91, 501–511 (2017).
Byron, A. et al. Glomerular cell cross-talk influences composition and assembly of extracellular matrix. J. Am. Soc. Nephrol. 25, 953 (2014).
Nagaraj, N. et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548 (2011).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Rinschen, M. M. et al. Quantitative deep mapping of the cultured podocyte proteome uncovers shifts in proteostatic mechanisms during differentiation. Am. J. Physiol. Cell Physiol. 311, C404–C417 (2016).
Boerries, M. et al. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 83, 1052–1064 (2013).
Rinschen, M. M. et al. A Multi-layered quantitative in vivo expression atlas of the podocyte unravels kidney disease candidate genes. Cell Rep. 23, 2495–2508 (2018).
Geyer, P. E., Holdt, L. M., Teupser, D. & Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 13, 942 (2017).
Martini, S., Eichinger, F., Nair, V. & Kretzler, M. Defining human diabetic nephropathy on the molecular level: integration of transcriptomic profiles with biological knowledge. Rev. Endocr. Metab. Disord. 9, 267–274 (2008).
Niewczas, M. A. et al. A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat. Med. 25, 805–813 (2019).
Pedigo, C. E. et al. Local TNF causes NFATc1-dependent cholesterol-mediated podocyte injury. J. Clin. Invest. 126, 3336–3350 (2016).
Nagaraj, N. & Mann, M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J. Proteome Res. 10, 637–645 (2011).
Hogan, M. C. et al. Identification of biomarkers for PKD1 using urinary exosomes. J. Am. Soc. Nephrol. 26, 1661–1670 (2015).
Salih, M. et al. Proteomics of urinary vesicles links plakins and complement to polycystic kidney disease. J. Am. Soc. Nephrol. 27, 3079–3092 (2016).
Diedrich, B. & Dengjel, J. Insights into autosomal dominant polycystic kidney disease by quantitative mass spectrometry-based proteomics. Cell Tissue Res. 369, 41–51 (2017).
Menezes, L. F. & Germino, G. G. Systems biology of polycystic kidney disease: a critical review. Wiley Interdiscip. Rev. Syst. Biol. Med. 7, 39–52 (2015).
Klein, J. et al. Fetal urinary peptides to predict postnatal outcome of renal disease in fetuses with posterior urethral valves (PUV). Sci. Transl Med. 5, 198ra106 (2013).
Klein, J. et al. Urinary peptidomics provides a noninvasive humanized readout of diabetic nephropathy in mice. Kidney Int. 90, 1045–1055 (2016).
Krochmal, M. et al. Urinary peptidomics analysis reveals proteases involved in diabetic nephropathy. Sci. Rep. 7, 15160 (2017).
Nkuipou-Kenfack, E. et al. Identification of ageing-associated naturally occurring peptides in human urine. Oncotarget 6, 34106–34117 (2015).
Merchant, M. L. et al. Urinary peptidome may predict renal function decline in type 1 diabetes and microalbuminuria. J. Am. Soc. Nephrol. 20, 2065–2074 (2009).
Van, J. A. D. et al. Peptidomic analysis of urine from youths with early type 1 diabetes reveals novel bioactivity of uromodulin peptides in vitro. Mol. Cell Proteom. 19, 501–517 (2020).
Tofte, N. et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomised placebo-controlled trial. Lancet Diabetes Endocrinol. 8, 301–312 (2020).
Krochmal, M., Schanstra, J. P. & Mischak, H. Urinary peptidomics in kidney disease and drug research. Expert Opin. Drug Discov. 13, 259–268 (2018).
Frantzi, M., Mischak, H. & Latosinska, A. Clinical proteomics on the path toward implementation: first promises delivered. Proteom. Clin. Appl. 13, e1800094 (2019).
Sethi, S. & Theis, J. D. Pathology and diagnosis of renal non-AL amyloidosis. J. Nephrol. 31, 343–350 (2017).
Sethi, S. et al. Apolipoprotein CII amyloidosis associated with p.Lys41Thr mutation. Kidney Int. Rep. 3, 1193–1201 (2018).
Qi, W. et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat. Med. 23, 753–762 (2017).
Slavov, N. Unpicking the proteome in single cells. Science 367, 512–513 (2020).
Koehler, S. et al. Proteome analysis of isolated podocytes reveals stress responses in glomerular sclerosis. J. Am. Soc. Nephrol. 31, 544–559 (2020).
Himmerkus, N. et al. Viewing cortical collecting duct function through phenotype-guided single-tubule proteomics. Function 1, zqaa007 (2020).
Limbutara, K., Chou, C.-L. & Knepper, M. A. Quantitative proteomics of All 14 renal tubule segments in rat. J. Am. Soc. Nephrol. 31, 1255–1266 (2020).
Rinschen, M. M., Limbutara, K., Knepper, M. A., Payne, D. M. & Pisitkun, T. From molecules to mechanisms: functional proteomics and its application to renal tubule physiology. Physiol. Rev. 98, 2571–2606 (2018).
Sung, C.-C. et al. RNA-Seq and protein mass spectrometry in microdissected kidney tubules reveal signaling processes initiating lithium-induced nephrogenic diabetes insipidus. Kidney Int. 96, 363–377 (2019).
Carr, S. A. et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteom. 13, 907–917 (2014).
Guo, T. et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat. Med. 21, 407–413 (2015).
Andeen, N. K., Yang, H.-Y., Dai, D.-F., MacCoss, M. J. & Smith, K. D. DnaJ homolog subfamily B member 9 is a putative autoantigen in fibrillary GN. J. Am. Soc. Nephrol. 29, 231–239 (2018).
Dasari, S. et al. DnaJ heat shock protein family B member 9 is a novel biomarker for fibrillary GN. J. Am. Soc. Nephrol. 29, 51–56 (2018).
Merchant, M. L. et al. Proteomic analysis identifies distinct glomerular extracellular matrix in collapsing focal segmental glomerulosclerosis. J. Am. Soc. Nephrol. 31, 1883–1904 (2020).
Hubner, N. C. & Mann, M. Extracting gene function from protein-protein interactions using quantitative BAC InteraCtomics (QUBIC). Methods 53, 453–459 (2011).
Kohli, P. et al. Label-free quantitative proteomic analysis of the YAP/TAZ interactome. Am. J. Physiol. Cell Physiol. 306, C805–C818 (2014).
Klein, J. B. Applying proteomics to detect early signs of chronic kidney disease: where has the magic gone? Expert. Rev. Proteom. 14, 387–390 (2017).
Francis, J. M., Beck, L. H. & Salant, D. J. Membranous nephropathy: a journey from bench to bedside. Am. J. Kidney Dis. 68, 138–147 (2016).
Tomas, N. M. et al. Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N. Engl. J. Med. 371, 2277–2287 (2014).
Sethi, S. et al. Exostosin 1/exostosin 2-associated membranous nephropathy. J. Am. Soc. Nephrol. 30, 1123–1136 (2019).
Laghmani, K. et al. Polyhydramnios, transient antenatal Bartter’s syndrome, and MAGED2 mutations. N. Engl. J. Med. 374, 1853–1863 (2016).
Legrand, A. et al. Prevalence of Novel MAGED2 Mutations in antenatal Bartter syndrome. Clin. J. Am. Soc. Nephrol. 13, 242–250 (2018).
Grahammer, F. et al. A flexible, multilayered protein scaffold maintains the slit in between glomerular podocytes. JCI Insight 1, e86177 (2016).
Rinschen, M. M. et al. The ubiquitin ligase Ubr4 controls stability of podocin/MEC-2 supercomplexes. Hum. Mol. Genet. 25, 1328–1344 (2016).
Anders, U. et al. SPRi-MALDI MS: characterization and identification of a kinase from cell lysate by specific interaction with different designed ankyrin repeat proteins. Anal. Bioanal. Chem. 409, 1827–1836 (2017).
Florinskaya, A. et al. SPR biosensors in direct molecular fishing: implications for protein interactomics. Sensors 18, 1616 (2018).
Kleifeld, O. et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 (2010).
Choudhary, C., Weinert, B. T., Nishida, Y., Verdin, E. & Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 (2014).
Macek, B., Mann, M. & Olsen, J. V. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 (2009).
Lienhard, G. E. Non-functional phosphorylations? Trends Biochem. Sci. 33, 351–352 (2008).
Landry, C. R., Levy, E. D. & Michnick, S. W. Weak functional constraints on phosphoproteomes. Trends Genet. 25, 193–197 (2009).
Ochoa, D. et al. The functional landscape of the human phosphoproteome. Nat. Biotechnol. 38, 365–373 (2020).
Beltrao, P. et al. Systematic functional prioritization of protein posttranslational modifications. Cell 150, 413–425 (2012).
Tonna, S. J. et al. NPHS2 variation in focal and segmental glomerulosclerosis. BMC Nephrol. 9, 13 (2008).
Rinschen, M. M. et al. Phosphoproteomic analysis reveals regulatory mechanisms at the kidney filtration barrier. J. Am. Soc. Nephrol. 25, 1509–1522 (2014).
Rinschen, M. M. et al. Comparative phosphoproteomic analysis of mammalian glomeruli reveals conserved podocin C-terminal phosphorylation as a determinant of slit diaphragm complex architecture. Proteomics 15, 1326–1331 (2015).
Buvall, L. et al. Synaptopodin is a coincidence detector of tyrosine versus serine/threonine phosphorylation for the modulation of Rho protein crosstalk in podocytes. J. Am. Soc. Nephrol. 28, 837–851 (2017).
Miller, M. L. et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 1, ra2 (2008).
Garg, P., Verma, R., Nihalani, D., Johnstone, D. B. & Holzman, L. B. Neph1 cooperates with nephrin to transduce a signal that induces actin polymerization. Mol. Cell. Biol. 27, 8698–8712 (2007).
Schroeter, C. B. et al. Protein half-life determines expression of proteostatic networks in podocyte differentiation. FASEB J. 32, 4696–4713 (2018).
Yaddanapudi, S. et al. CD2AP in mouse and human podocytes controls a proteolytic program that regulates cytoskeletal structure and cellular survival. J. Clin. Invest. 121, 3965–3980 (2011).
Yamamoto-Nonaka, K. et al. Cathepsin D in podocytes is important in the pathogenesis of proteinuria and CKD. J. Am. Soc. Nephrol. 27, 2685–2700 (2016).
Garsen, M. et al. Cathepsin L is crucial for the development of early experimental diabetic nephropathy. Kidney Int. 90, 1012–1022 (2016).
Rinschen, M. M. et al. N-degradomic analysis reveals a proteolytic network processing the podocyte cytoskeleton. J. Am. Soc. Nephrol. 28, 2867–2878 (2017).
Rinschen, M. M., Huesgen, P. F. & Koch, R. E. The podocyte protease web: uncovering the gatekeepers of glomerular disease. Am. J. Physiol. Ren. Physiol. 315, F1812–F1816 (2018).
Späth, M. R. et al. The proteome microenvironment determines the protective effect of preconditioning in cisplatin-induced acute kidney injury. Kidney Int. 95, 333–349 (2019).
Bensimon, A., Heck, A. J. R. & Aebersold, R. Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 81, 379–405 (2012).
Creixell, P. et al. Pathway and network analysis of cancer genomes. Nat. Methods 12, 615–621 (2015).
Türei, D., Korcsmáros, T. & Saez-Rodriguez, J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 13, 966–967 (2016).
Rodchenkov, I. et al. Pathway commons 2019 update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 48, D489–D497 (2020).
Huang, J. K. et al. Systematic evaluation of molecular networks for discovery of disease genes. Cell Syst. 6, 484–495.e5 (2018).
Gonzalez-Vicente, A., Hopfer, U. & Garvin, J. L. Developing tools for analysis of renal genomic data: an invitation to participate. J. Am. Soc. Nephrol. 28, 3438–3440 (2017).
Terfve, C. et al. System-wide quantitative proteomics of the metabolic syndrome in mice: genotypic and dietary effects. J. Proteome Res. 16, 831–841 (2017).
Goh, W. W. B. & Wong, L. Networks in proteomics analysis of cancer. Curr. Opin. Biotechnol. 24, 1122–1128 (2013).
Schoeberl, B. et al. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci. Signal. 2, ra31 (2009).
Schoeberl, B. et al. Systems biology driving drug development: from design to the clinical testing of the anti-ErbB3 antibody seribantumab (MM-121). NPJ Syst. Biol. Appl. 3, 16034 (2017).
Kholodenko, B., Yaffe, M. B. & Kolch, W. Computational approaches for analyzing information flow in biological networks. Sci. Signal. 5, re1 (2012).
Le Novère, N. Quantitative and logic modelling of molecular and gene networks. Nat. Rev. Genet. 16, 146–158 (2015).
Lamb, J. et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17 (2017).
Williams, V. R. et al. Connectivity mapping of a chronic kidney disease progression signature identified lysine deacetylases as novel therapeutic targets. Kidney Int. 98, 116–132 (2020).
Tajti, F. et al. A functional landscape of CKD entities from public transcriptomic data. Kidney Int. Rep. 5, 211–224 (2020).
Schanstra, J. P. et al. Systems biology identifies cytosolic PLA2 as a target in vascular calcification treatment. JCI Insight 4, e125638 (2019).
Litichevskiy, L. et al. A library of phosphoproteomic and chromatin signatures for characterizing cellular responses to drug perturbations. Cell Syst. 6, 424–443.e7 (2018).
Calizo, R. C. et al. Disruption of podocyte cytoskeletal biomechanics by dasatinib leads to nephrotoxicity. Nat. Commun. 10, 2061 (2019).
Azeloglu, E. U. et al. Interconnected network motifs control podocyte morphology and kidney function. Sci. Signal. 7, ra12 (2014).
Moraru, I. I. et al. The virtual cell modeling and simulation software environment. IET Syst. Biol. 2, 352–362 (2008).
Falkenberg, C. V. et al. Fragility of foot process morphology in kidney podocytes arises from chaotic spatial propagation of cytoskeletal instability. PLoS Comput. Biol. 13, e1005433 (2017).
Rinschen, M. M. et al. YAP-mediated mechanotransduction determines the podocyte’s response to damage. Sci. Signal. 10, eaaf8165 (2017).
Wong, J. S., Meliambro, K., Ray, J. & Campbell, K. N. Hippo signaling in the kidney: the good and the bad. Am. J. Physiol. Ren. Physiol. 311, F241–F248 (2016).
Klein, J. et al. The KUPKB: a novel Web application to access multiomics data on kidney disease. FASEB J. 26, 2145–2153 (2012).
Rhodes, D. R. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180 (2007).
Vizcaíno, J. A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Sullivan, K. M. & Susztak, K. Unravelling the complex genetics of common kidney diseases: from variants to mechanisms. Nat. Rev. Nephrol. https://doi.org/10.1038/s41581-020-0298-1 (2020).
Gillies, C. E. et al. An eQTL landscape of kidney tissue in human nephrotic syndrome. Am. J. Hum. Genet. 103, 232–244 (2018).
Qiu, C. et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 24, 1721–1731 (2018).
Rinschen, M. M., Ivanisevic, J., Giera, M. & Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 20, 353–367 (2019).
Narita, T., Weinert, B. T. & Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174 (2019).
James, A. M. et al. The causes and consequences of nonenzymatic protein acylation. Trends Biochem. Sci. 43, 921–932 (2018).
Hart, G. W., Slawson, C., Ramirez-Correa, G. & Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 (2011).
Rinschen, M. M. et al. Metabolic rewiring of the hypertensive kidney. Sci. Signal. 12, eaax9760 (2019).
Terfve, C. D. A., Wilkes, E. H., Casado, P., Cutillas, P. R. & Saez-Rodriguez, J. Large-scale models of signal propagation in human cells derived from discovery phosphoproteomic data. Nat. Commun. 6, 1–11 (2015).
Kang, H. M. et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 21, 37–46 (2015).
Gordin, D. et al. Characterization of glycolytic enzymes and pyruvate kinase M2 in type 1 and 2 diabetic nephropathy. Diabetes Care 42, 1263–1273 (2019).
Tape, C. J. Systems biology analysis of heterocellular signaling. Trends Biotechnol. 34, 627–637 (2016).
Malone, A. F., Wu, H. & Humphreys, B. D. Bringing renal biopsy interpretation into the molecular age with single-cell RNA sequencing. Semin. Nephrol. 38, 31–39 (2018).
Höhne, M. et al. Single-nephron proteomes connect morphology and function in proteinuric kidney disease. Kidney Int. 93, 1308–1319 (2018).
Fribourg, M. et al. T-cell exhaustion correlates with improved outcomes in kidney transplant recipients. Kidney Int. 96, 436–449 (2019).
Brähler, S. et al. Opposing roles of dendritic cell subsets in experimental GN. J. Am. Soc. Nephrol. 29, 138–154 (2018).
Singh, N. et al. Development of a 2-dimensional atlas of the human kidney with imaging mass cytometry. JCI Insight 4, e129477 (2019).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Drewes, G. & Knapp, S. Chemoproteomics and chemical probes for target discovery. Trends Biotechnol. 36, 1275–1286 (2018).
Klaeger, S. et al. The target landscape of clinical kinase drugs. Science 358, eaan4368 (2017).
Huber, K. V. M. et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 508, 222–227 (2014).
Becher, I. et al. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat. Chem. Biol. 12, 908–910 (2016).
Bartram, M. P. et al. Three-layered proteomic characterization of a novel ACTN4 mutation unravels its pathogenic potential in FSGS. Hum. Mol. Genet. 25, 1152–1164 (2016).
Gillet, L. C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell Proteomics 11, O111.016717 (2012).
Ong, S.-E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteom. 1, 376–386 (2002).
McAlister, G. C. et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 84, 7469–7478 (2012).
Ngo, D. et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation 134, 270–285 (2016).
Knepper, M. A. Proteomics and the kidney. J. Am. Soc. Nephrol. 13, 1398–1408 (2002).
Hoppe-Seyler, F. & Butz, K. Peptide aptamers: powerful new tools for molecular medicine. J. Mol. Med. 78, 426–430 (2000).
Geyer, C. R., Colman-Lerner, A. & Brent, R. “Mutagenesis” by peptide aptamers identifies genetic network members and pathway connections. Proc. Natl Acad. Sci. USA 96, 8567–8572 (1999).
Anderson, N. L. et al. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA). J. Proteome Res. 3, 235–244 (2004).
Neubert, H. et al. 2018 white paper on recent issues in bioanalysis: focus on immunogenicity assays by hybrid LBA/LCMS and regulatory feedback (Part 2 — PK, PD & ADA assays by hybrid LBA/LCMS & regulatory agencies’ inputs on bioanalysis, biomarkers and immunogenicity). Bioanalysis 10, 1897–1917 (2018).
Bodenmiller, B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst. 2, 225–238 (2016).
Casadonte, R. et al. Imaging mass spectrometry analysis of renal amyloidosis biopsies reveals protein co-localization with amyloid deposits. Anal. Bioanal. Chem. 407, 5323–5331 (2015).
Rhee, H.-W. et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013).
Acknowledgements
The authors apologize to all researchers whose work could not be cited due to space limitations. M.M.R. was supported by the DFG (RI2811/1 and RI2811/2), as well as the Young Investigator Award from the Novo Nordisk Foundation, grant number NNF19OC0056043. The authors thank Nicolas Palacio-Escat (Heidelberg University) for help with the initial figure panels and Aurelien Dugourd (Aachen University Hospital) for critically reading the manuscript before submission.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, made substantial contributions to discussions of the content, wrote the article, and reviewed or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Nephrology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Connectivity map: https://clue.io/cmap
Proteome central: http://proteomecentral.proteomexchange.org/cgi/GetDataset
Renal genomics portal: https://www.wikipathways.org/index.php/Portal:RenalGenomics
Glossary
- Proteoforms
-
Distinct forms of a protein molecule that arise from the same transcript or gene.
- Intensity-based absolute quantification
-
An algorithm used for protein copy number estimation in which total protein intensity (that is, the sum of all peptide intensities) is divided by the number of peptides.
- Surface plasmon resonance
-
A biophysical method used to probe protein–protein (and molecule–molecule) interactions.
- PhoNEMES modelling
-
A computational tool used to build logic models of signalling networks from discovery mass-spectrometry-based phosphoproteomic data.
Rights and permissions
About this article
Cite this article
Rinschen, M.M., Saez-Rodriguez, J. The tissue proteome in the multi-omic landscape of kidney disease. Nat Rev Nephrol 17, 205–219 (2021). https://doi.org/10.1038/s41581-020-00348-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41581-020-00348-5
This article is cited by
-
Proteomic profiling of cerebrospinal fluid in pediatric myelin oligodendrocyte glycoprotein antibody-associated disease
World Journal of Pediatrics (2024)
-
Metabolome panels as potential noninvasive biomarkers for primary glomerulonephritis sub-types: meta-analysis of profiling metabolomics studies
Scientific Reports (2023)
-
Intrarenal 1-methoxypyrene, an aryl hydrocarbon receptor agonist, mediates progressive tubulointerstitial fibrosis in mice
Acta Pharmacologica Sinica (2022)
-
Non-invasive molecular imaging of kidney diseases
Nature Reviews Nephrology (2021)
-
Proteolysis and inflammation of the kidney glomerulus
Cell and Tissue Research (2021)