Abstract
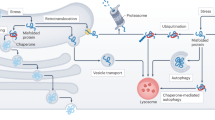
Ageing is a major risk factor for the development of many diseases, prominently including neurodegenerative disorders such as Alzheimer disease and Parkinson disease. A hallmark of many age-related diseases is the dysfunction in protein homeostasis (proteostasis), leading to the accumulation of protein aggregates. In healthy cells, a complex proteostasis network, comprising molecular chaperones and proteolytic machineries and their regulators, operates to ensure the maintenance of proteostasis. These factors coordinate protein synthesis with polypeptide folding, the conservation of protein conformation and protein degradation. However, sustaining proteome balance is a challenging task in the face of various external and endogenous stresses that accumulate during ageing. These stresses lead to the decline of proteostasis network capacity and proteome integrity. The resulting accumulation of misfolded and aggregated proteins affects, in particular, postmitotic cell types such as neurons, manifesting in disease. Recent analyses of proteome-wide changes that occur during ageing inform strategies to improve proteostasis. The possibilities of pharmacological augmentation of the capacity of proteostasis networks hold great promise for delaying the onset of age-related pathologies associated with proteome deterioration and for extending healthspan.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008). This paper presents the first introduction of the term proteostasis and of the proteostasis concept.
Klaips, C. L., Jayaraj, G. G. & Hartl, F. U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217, 51–63 (2017).
Taylor, R. C. & Dillin, A. Aging as an event of proteostasis collapse. Cold Spring Harb. Persp. Biol. 3, a004440 (2011).
Labbadia, J. & Morimoto, R. I. The biology of proteostasis inaging and disease. Annu. Rev. Biochem. 84, 435–464 (2015).
Picotti, P. et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 494, 266–270 (2013).
Kulak, N. A., Geyer, P. E. & Mann, M. Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol. Cell. Proteomics 16, 694–705 (2017).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Boucher, J. I., Bolon, D. N. & Tawfik, D. S. Quantifying and understanding the fitness effects of protein mutations: laboratory versus nature. Protein Sci. 25, 1219–1226 (2016).
Kundra, R., Ciryam, P., Morimoto, R. I., Dobson, C. M. & Vendruscolo, M. Protein homeostasis of a metastable subproteome associated with Alzheimer’s disease. Proc. Natl Acad. Sci. USA 114, E5703–E5711 (2017).
Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R. & Morimoto, R. I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 (2006). This paper demonstrates that expression of disease-related mutant proteins disrupts global protein folding.
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Geiger, T., Wehner, A., Schaab, C., Cox, J. & Mann, M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050 (2012).
Demarest, S. J. et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415, 549–553 (2002).
Dunker, A. K., Silman, I., Uversky, V. N. & Sussman, J. L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18, 756–764 (2008).
Hartl, F. U. Molecular chaperones in cellular protein folding. Nature 381, 571–579 (1996).
Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M. & Hartl, F. U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 (2013).
Hartl, F. U., Bracher, A. & Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011).
Balchin, D., Hayer-Hartl, M. & Hartl, F. U. In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016).
Carra, S. et al. The growing world of small heat shock proteins: from structure to functions. Cell Stress Chaperones 22, 601–611 (2017).
Lee, C., Kim, H. & Bardwell, J. C. A. Electrostatic interactions are important for chaperone-client interaction in vivo. Microbiology 164, 992–997 (2018).
Joachimiak, L. A., Walzthoeni, T., Liu, C. W., Aebersold, R. & Frydman, J. The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159, 1042–1055 (2014).
Koldewey, P., Stull, F., Horowitz, S., Martin, R. & Bardwell, J. C. A. Forces driving chaperone action. Cell 166, 369–379 (2016).
Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017).
Young, J. C., Hoogenraad, N. J. & Hartl, F. U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 (2003).
Liu, Q., D’Silva, P., Walter, W., Marszalek, J. & Craig, E. A. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300, 139–141 (2003).
Schneider, H. C. et al. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371, 768–774 (1994).
Zou, J., Guo, Y., Guettouche, T., Smith, D. F. & Voellmy, R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471–480 (1998).
Zheng, X. et al. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 5, e18638 (2016).
Anckar, J. & Sistonen, L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80, 1089–1115 (2011).
Gomez-Pastor, R., Burchfiel, E. T. & Thiele, D. J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19 (2018).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Shpilka, T. & Haynes, C. M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19, 109–120 (2018).
Wyatt, A. R., Yerbury, J. J., Ecroyd, H. & Wilson, M. R. Extracellular chaperones and proteostasis. Annu. Rev. Biochem. 82, 295–322 (2013).
Glotzer, M., Murray, A. W. & Kirschner, M. W. Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138 (1991).
Faust, J. R., Luskey, K. L., Chin, D. J., Goldstein, J. L. & Brown, M. S. Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc. Natl Acad. Sci. USA 79, 5205–5209 (1982).
Murakami, Y. et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599 (1992).
Ciechanover, A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Best Pract. Res. Clin. Haematol. 30, 341–355 (2017).
Dikic, I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 (2017).
Varshavsky, A. The ubiquitin system, an immense realm. Annu. Rev. Biochem. 81, 167–176 (2012).
Arndt, V., Rogon, C. & Höhfeld, J. To be, or not to be — molecular chaperones in protein degradation. Cell. Mol. Life Sci. 64, 2525–2541 (2007).
Shiber, A. & Ravid, T. Chaperoning proteins for destruction: diverse roles of Hsp70 chaperones and their co-chaperones in targeting misfolded proteins to the proteasome. Biomolecules 4, 704–724 (2014).
Tekirdag, K. & Cuervo, A. M. Chaperone-mediated autophagy and endosomal microautophagy: joint by a chaperone. J. Biol. Chem. 293, 5414–5424 (2018).
Esser, C., Alberti, S. & Hohfeld, J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta 1695, 171–188 (2004).
Rosser, M. F., Washburn, E., Muchowski, P. J., Patterson, C. & Cyr, D. M. Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 282, 22267–22277 (2007).
Rosenbaum, J. C. et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell 41, 93–106 (2011).
Yanagitani, K., Juszkiewicz, S. & Hegde, R. S. UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357, 472–475 (2017).
Hwang, C. S., Shemorry, A. & Varshavsky, A. N-Terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010). This paper introduces amino-terminal acetylation as a signal for proteasomal degradation to regulate the removal of nonassembled protein subunits of oligomeric complexes.
Kettern, N., Dreiseidler, M., Tawo, R. & Hohfeld, J. Chaperone-assisted degradation: multiple paths to destruction. Biol. Chem. 391, 481–489 (2010).
Arndt, V. et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143–148 (2010).
Gamerdinger, M., Kaya, A. M., Wolfrum, U., Clement, A. M. & Behl, C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 12, 149–156 (2011).
Chiang, H. L., Terlecky, S. R., Plant, C. P. & Dice, J. F. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382–385 (1989).
Kaushik, S. & Cuervo, A. M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018).
Sahu, R. et al. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20, 131–139 (2011).
Sies, H., Berndt, C. & Jones, D. P. Oxidative stress. Annu. Rev. Biochem. 86, 715–748 (2017).
Jacobson, T. et al. Cadmium causes misfolding and aggregation of cytosolic proteins in yeast. Mol. Cell. Biol. 37, e00490–16 (2017).
Lang, L., Kurnik, M., Danielsson, J. & Oliveberg, M. Fibrillation precursor of superoxide dismutase 1 revealed by gradual tuning of the protein-folding equilibrium. Proc. Natl Acad. Sci. USA 109, 17868–17873 (2012).
Schubert, U. et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 (2000).
Vabulas, R. M. & Hartl, F. U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310, 1960–1963 (2005).
Duttler, S., Pechmann, S. & Frydman, J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 (2013).
Ward, C. L. & Kopito, R. R. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J. Biol. Chem. 269, 25710–25718 (1994).
Lukacs, G. L. et al. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 13, 6076–6086 (1994).
Bruns, C. K. & Kopito, R. R. Impaired post-translational folding of familial ALS-linked Cu, Zn superoxide dismutase mutants. EMBO J. 26, 855–866 (2007).
Brandman, O. & Hegde, R. S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23, 7–15 (2016).
Choe, Y. J. et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 (2016).
Yonashiro, R. et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife 5, e11794 (2016).
Defenouillere, Q. et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl Acad. Sci. USA 110, 5046–5051 (2013).
Chu, J. et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl Acad. Sci. USA 106, 2097–2103 (2009).
Nedialkova, D. D. & Leidel, S. A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 (2015). This paper shows that codon-specific translational pausing can cause protein misfolding.
Wrobel, L. et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488 (2015).
Park, S. H. et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell 18, 153–165 (2007).
Alzheimer, A., Förstl, H. & Levy, R. On certain peculiar diseases of old age. Hist. Psychiatry 2, 74–101 (1991).
Mukherjee, A., Morales-Scheihing, D., Butler, P. C. & Soto, C. Type 2 diabetes as a protein misfolding disease. Trends Mol. Med. 21, 439–449 (2015).
Iadanza, M. G., Jackson, M. P., Hewitt, E. W., Ranson, N. A. & Radford, S. E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 19, 755–773 (2018).
Winklhofer, K. F., Tatzelt, J. & Haass, C. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 27, 336–349 (2008).
Lukacs, G. L. & Verkman, A. S. CFTR: folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol. Med. 18, 81–91 (2012).
Riek, R. & Eisenberg, D. S. The activities of amyloids from a structural perspective. Nature 539, 227–235 (2016).
Landreh, M. et al. The formation, function and regulation of amyloids: insights from structural biology. J. Intern. Med. 280, 164–176 (2016).
Chiti, F. & Dobson, C. M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017).
Tipping, K. W., van Oosten-Hawle, P., Hewitt, E. W. & Radford, S. E. Amyloid fibres: inert end-stage aggregates or key players in disease? Trends Biochem. Sci. 40, 719–727 (2015).
Arosio, P., Knowles, T. P. & Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 17, 7606–7618 (2015).
Arosio, P., Vendruscolo, M., Dobson, C. M. & Knowles, T. P. Chemical kinetics for drug discovery to combat protein aggregation diseases. Trends Pharmacol. Sci. 35, 127–135 (2014).
Wagner, A. S. et al. Self-assembly of mutant huntingtin exon-1 fragments into large complex fibrillar structures involves nucleated branching. J. Mol. Biol. 430, 1725–1744 (2018).
Brundin, P., Melki, R. & Kopito, R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat. Rev. Mol. Cell Biol. 11, 301–307 (2010).
Li, J. Y. et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat. Med. 14, 501–503 (2008).
Sibilla, C. & Bertolotti, A. Prion properties of SOD1 in amyotrophic lateral sclerosis and potential therapy. Cold Spring Harb. Persp. Biol. 9, a024141 (2017).
Guo, J. L. et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 213, 2635–2654 (2016).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Chapman, M. R. et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 (2002).
Fowler, D. M. et al. Functional amyloid formation within mammalian tissue. PLOS Biol. 4, e6 (2006).
Sengupta, U., Nilson, A. N. & Kayed, R. The role of amyloid-beta oligomers in toxicity, propagation, and immunotherapy. EBioMedicine 6, 42–49 (2016).
Jackson, M. P. & Hewitt, E. W. Why are functional amyloids non-toxic in humans? Biomolecules 7, 71 (2017).
Kayed, R. et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
Miller, J. et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol. 7, 925–934 (2011).
Cheon, M. et al. Structural reorganisation and potential toxicity of oligomeric species formed during the assembly of amyloid fibrils. PLOS Comput. Biol. 3, 1727–1738 (2007).
Sangwan, S. et al. Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc. Natl Acad. Sci. USA 114, 8770–8775 (2017).
Kim, Y. E. et al. Soluble oligomers of PolyQ-expanded huntingtin target a multiplicity of key cellular factors. Mol. Cell 63, 951–964 (2016).
Franzmann, T. M. et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654 (2018).
Mateju, D. et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687 (2017).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Gopal, P. P., Nirschl, J. J., Klinman, E. & Holzbaur, E. L. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc. Natl Acad. Sci. USA 114, E2466–E2475 (2017).
Alberti, S. & Hyman, A. A. Are aberrant phase transitions a driver of cellular aging? Bioessays 38, 959–968 (2016).
Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T. & Lansbury, P. T. Jr. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418, 291 (2002). This paper demonstrates that mutant amyloidogenic proteins can form pores in membranes.
Anguiano, M., Nowak, R. J. & Lansbury, P. T. Jr. Protofibrillar islet amyloid polypeptide permeabilizes synthetic vesicles by a pore-like mechanism that may be relevant to type II diabetes. Biochemistry 41, 11338–11343 (2002).
Lashuel, H. A. & Lansbury, P. T. Jr. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 39, 167–201 (2006).
Milanesi, L. et al. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc. Natl Acad. Sci. USA 109, 20455–20460 (2012). This paper demonstrates use of cryo-electron tomography to show that fibrils can damage membranes.
Bauerlein, F. J. B. et al. In situ architecture and cellular interactions of PolyQ inclusions. Cell 171, 179–187 (2017).
Chou, C. C. et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239 (2018).
Woerner, A. C. et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351, 173–176 (2016).
Zhang, Y. J. et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 19, 668–677 (2016).
Gasset-Rosa, F. et al. Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94, 48–57 (2017).
Ramdzan, Y. M. et al. Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep. 19, 919–927 (2017).
Olzscha, H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 (2011). This paper describes soluble protein sequestration in aggregates as a basic mechanism of the toxicity of aggregates.
Lin, Y. et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802 (2016).
Lee, K. H. et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788 (2016).
Hosp, F. et al. Spatiotemporal proteomic profiling of huntington’s disease inclusions reveals widespread loss of protein function. Cell Rep. 21, 2291–2303 (2017).
Park, S. H. et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145 (2013).
Yu, A. et al. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc. Natl Acad. Sci. USA 111, E1481–E1490 (2014).
Chafekar, S. M. & Duennwald, M. L. Impaired heat shock response in cells expressing full-length polyglutamine-expanded huntingtin. PLOS ONE 7, e37929 (2012).
Roth, D. M. et al. Modulation of the maladaptive stress response to manage diseases of protein folding. PLOS Biol. 12, e1001998 (2014).
Ashkenazi, A. et al. Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 108–111 (2017).
Bence, N. F., Sampat, R. M. & Kopito, R. R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552–1555 (2001). This paper shows that expression of aggregation-prone proteins interferes with the function of the UPS.
Hipp, M. S. et al. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington’s disease. J. Cell Biol. 196, 573–587 (2012).
Guo, Q. et al. In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell 172, 696–705 (2018). This paper presents uses of cryo-electron tomography to show that proteasomes are sequestered inside aggregates.
Deriziotis, P. et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 30, 3065–3077 (2011).
Hipp, M. S., Park, S. H. & Hartl, F. U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 (2014).
Behrends, C. et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell 23, 887–897 (2006).
Muchowski, P. J. et al. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl Acad. Sci. USA 97, 7841–7846 (2000).
Schaffar, G. et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell 15, 95–105 (2004).
Dedmon, M. M., Christodoulou, J., Wilson, M. R. & Dobson, C. M. Heat shock protein 70 inhibits alpha-synuclein fibril formation via preferential binding to prefibrillar species. J. Biol. Chem. 280, 14733–14740 (2005).
Rujano, M. A., Kampinga, H. H. & Salomons, F. A. Modulation of polyglutamine inclusion formation by the Hsp70 chaperone machine. Exp. Cell Res. 313, 3568–3578 (2007).
Kakkar, V., Kuiper, E. F., Pandey, A., Braakman, I. & Kampinga, H. H. Versatile members of the DNAJ family show Hsp70 dependent anti-aggregation activity on RING1 mutant parkin C289G. Sci. Rep. 6, 34830 (2016).
Lotz, G. P. et al. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J. Biol. Chem. 285, 38183–38193 (2010).
Tam, S., Geller, R., Spiess, C. & Frydman, J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 8, 1155–1162 (2006).
Kitamura, A. et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 8, 1163–1170 (2006).
Cohen, S. I. A. et al. A molecular chaperone breaks the catalytic cycle that generates toxic Abeta oligomers. Nat. Struct. Mol. Biol. 22, 207–213 (2015).
Kordasiewicz, H. B. et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044 (2012).
Yamamoto, A., Lucas, J. J. & Hen, R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell 101, 57–66 (2000). This paper demonstrates that disease-related aggregates can be cleared when synthesis of the disease protein is blocked.
Glover, J. R. & Lindquist, S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998).
Parsell, D. A., Kowal, A. S., Singer, M. A. & Lindquist, S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372, 475–478 (1994).
Nillegoda, N. B. et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524, 247–251 (2015). This paper describes a metazoan chaperone system for protein disaggregation.
Mogk, A., Bukau, B. & Kampinga, H. H. Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226 (2018).
Escusa-Toret, S., Vonk, W. I. & Frydman, J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15, 1231–1243 (2013). This paper defines aggregation of misfolded proteins as a process important in maintaining proteostasis during stress.
Miller, S. B. et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34, 778–797 (2015).
Wallace, E. W. et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 (2015).
Malinovska, L., Kroschwald, S., Munder, M. C., Richter, D. & Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 23, 3041–3056 (2012).
Grousl, T. et al. A prion-like domain in Hsp42 drives chaperone-facilitated aggregation of misfolded proteins. J. Cell Biol. 217, 1269–1285 (2018).
Mogk, A. & Bukau, B. Role of sHsps in organizing cytosolic protein aggregation and disaggregation. Cell Stress Chaperones 22, 493–502 (2017).
Kaganovich, D., Kopito, R. & Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 (2008). This paper introduces the concept of spatial control of aggregate deposition in distinct cellular locations.
Kopito, R. R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (2000).
Arrasate, M., Mitra, S., Schweitzer, E., Segal, M. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004). This paper shows that inclusion-body formation can be beneficial by sequestering toxic aggregates.
Liu, B. et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140, 257–267 (2010). This paper describes the machinery that controls the asymmetric distribution of aggregates during cell division in budding yeast.
Hill, S. M., Hanzen, S. & Nystrom, T. Restricted access: spatial sequestration of damaged proteins during stress and aging. EMBO Rep. 18, 377–391 (2017).
Sontag, E. M., Samant, R. S. & Frydman, J. Mechanisms and functions of spatial protein quality control. Annu. Rev. Biochem. 86, 97–122 (2017).
Ruan, L. et al. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446 (2017).
Rousseau, E. et al. Targeting expression of expanded polyglutamine proteins to the endoplasmic reticulum or mitochondria prevents their aggregation. Proc. Natl Acad. Sci. USA 101, 9648–9653 (2004).
Vincenz-Donnelly, L. et al. High capacity of the endoplasmic reticulum to prevent secretion and aggregation of amyloidogenic proteins. EMBO J. 37, 337–350 (2018).
Rubinsztein, D. C., Marino, G. & Kroemer, G. Autophagy and aging. Cell 146, 682–695 (2011).
Min, J. N. et al. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 28, 4018–4025 (2008).
Labbadia, J. & Morimoto, R. I. Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Mol. Cell 59, 639–650 (2015).
David, D. C. et al. Widespread protein aggregation as an inherent part of aging in C. elegans. PLOS Biol. 8, e1000450 (2010).
Reis-Rodrigues, P. et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell 11, 120–127 (2012).
Liang, V. et al. Altered proteostasis in aging and heat shock response in C. elegans revealed by analysis of the global and de novo synthesized proteome. Cell. Mol. Life Sci. 71, 3339–3361 (2014).
Walther, D. M. et al. Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919–932 (2015). This paper presents an analysis of proteome changes during the lifespan of C. elegans.
Zimmerman, S. M., Hinkson, I. V., Elias, J. E. & Kim, S. K. Reproductive aging drives protein accumulation in the uterus and limits lifespan in C. elegans. PLOS Genet. 11, e1005725 (2015).
Waldera-Lupa, D. M. et al. Proteome-wide analysis reveals an age-associated cellular phenotype of in situ aged human fibroblasts. Aging 6, 856–878 (2014).
Walther, D. M. & Mann, M. Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol. Cell. Proteomics 10, M110.004523 (2011).
Ori, A. et al. Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 1, 224–237 (2015).
Ciryam, P., Kundra, R., Morimoto, R. I., Dobson, C. M. & Vendruscolo, M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 36, 72–77 (2015).
Ciryam, P., Tartaglia, G. G., Morimoto, R. I., Dobson, C. M. & Vendruscolo, M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 5, 781–790 (2013). This paper introduces supersaturation as a concept in controlling protein solubility.
Powers, E. T., Morimoto, R. I., Dillin, A., Kelly, J. W. & Balch, W. E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991 (2009).
Sala, A. J., Bott, L. C. & Morimoto, R. I. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 216, 1231–1241 (2017).
Brehme, M. et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 9, 1135–1150 (2014). This paper presents a census of the human chaperome and a description of a subnetwork that safeguards proteostasis.
Kitamura, A. et al. Dysregulation of the proteasome increases the toxicity of ALS-linked mutant SOD1. Genes Cells 19, 209–224 (2014).
Gupta, R. et al. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods 8, 879–884 (2011).
Vilchez, D. et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489, 304–308 (2012). This paper shows that stem cells have an increased level of proteasomal activity that is regulated by the proteasome subunit PSMD11.
Noormohammadi, A. et al. Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat. Commun. 7, 13649 (2016).
Bufalino, M. R., DeVeale, B. & van der Kooy, D. The asymmetric segregation of damaged proteins is stem cell-type dependent. J. Cell Biol. 201, 523–530 (2013).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Moore, D. L., Pilz, G. A., Arauzo-Bravo, M. J., Barral, Y. & Jessberger, S. A mechanism for the segregation of age in mammalian neural stem cells. Science 349, 1334–1338 (2015).
Kenyon, C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Phil. Trans. R. Soc. B 366, 9–16 (2011).
Kenyon, C., Chang, J., Gensch, E., Rudner, A. & Tabtiang, R. A. C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 (1993). This paper shows that mutations in the daf-2 gene cause a dramatic lifespan extension in C. elegans.
Kirstein-Miles, J., Scior, A., Deuerling, E. & Morimoto, R. I. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 32, 1451–1468 (2013).
Stout, G. J. et al. Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism. Mol. Syst. Biol. 9, 679 (2013).
Tsaytler, P., Harding, H. P., Ron, D. & Bertolotti, A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332, 91–94 (2011).
Frakes, A. E. & Dillin, A. The UPR(ER): sensor and coordinator of organismal homeostasis. Mol. Cell 66, 761–771 (2017).
Hansen, M. et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110 (2007).
Pan, K. Z. et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119 (2007).
Syntichaki, P., Troulinaki, K. & Tavernarakis, N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 445, 922–926 (2007).
Sherman, M. Y. & Qian, S. B. Less is more: improving proteostasis by translation slow down. Trends Biochem. Sci. 38, 585–591 (2013).
Vilchez, D., Saez, I. & Dillin, A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 5, 5659 (2014).
Caniard, A. et al. Proteasome function is not impaired in healthy aging of the lung. Aging 7, 776–792 (2015).
Hamer, G., Matilainen, O. & Holmberg, C. I. A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat. Methods 7, 473–478 (2010).
Tsakiri, E. N. et al. Differential regulation of proteasome functionality in reproductive versus somatic tissues of Drosophila during aging or oxidative stress. FASEB J. 27, 2407–2420 (2013).
Morrow, G. & Tanguay, R. M. Drosophila melanogaster Hsp22: a mitochondrial small heat shock protein influencing the aging process. Front. Genet. 6, 1026 (2015).
Yamaguchi, T. et al. Age-related increase of insoluble, phosphorylated small heat shock proteins in human skeletal muscle. J. Gerontol. A 62, 481–489 (2007).
Jiao, W., Li, P., Zhang, J., Zhang, H. & Chang, Z. Small heat-shock proteins function in the insoluble protein complex. Biochem. Biophys. Res. Commun. 335, 227–231 (2005).
Khan, S., Khamis, I. & Heikkila, J. J. The small heat shock protein, HSP30, is associated with aggresome-like inclusion bodies in proteasomal inhibitor-, arsenite-, and cadmium-treated Xenopus kidney cells. Comp. Biochem. Physiol. A 189, 130–140 (2015).
Walker, G. A. & Lithgow, G. J. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2, 131–139 (2003).
Cohen, E., Bieschke, J., Perciavalle, R. M., Kelly, J. W. & Dillin, A. Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006). This paper provides evidence for controlled protein aggregation during ageing as a beneficial process.
El-Ami, T. et al. A novel inhibitor of the insulin/IGF signaling pathway protects from age-onset, neurodegeneration-linked proteotoxicity. Aging Cell 13, 165–174 (2014).
Moll, L., Ben-Gedalya, T., Reuveni, H. & Cohen, E. The inhibition of IGF-1 signaling promotes proteostasis by enhancing protein aggregation and deposition. FASEB J. 30, 1656–1669 (2016).
Wild, E. J. & Tabrizi, S. J. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 16, 837–847 (2017).
Cohen, F. E. & Kelly, J. W. Therapeutic approaches to protein-misfolding diseases. Nature 426, 905–909 (2003).
Sevigny, J. et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 537, 50–56 (2016).
Bulawa, C. E. et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl Acad. Sci. USA 109, 9629–9634 (2012). This paper describes the first clinically effective anti-aggregation drug.
Baranczak, A. & Kelly, J. W. A current pharmacologic agent versus the promise of next generation therapeutics to ameliorate protein misfolding and/or aggregation diseases. Curr. Opin. Chem. Biol. 32, 10–21 (2016).
Paxman, R. et al. Pharmacologic ATF6 activating compounds are metabolically activated to selectively modify endoplasmic reticulum proteins. eLife 7, e37168 (2018).
Krobitsch, S. & Lindquist, S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl Acad. Sci. USA 97, 1589–1594 (2000).
Fonte, V. et al. Suppression of in vivo beta-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J. Biol. Chem. 283, 784–791 (2008).
Auluck, P. K., Chan, H. Y., Trojanowski, J. Q., Lee, V. M. & Bonini, N. M. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295, 865–868 (2002).
Warrick, J. M. et al. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23, 425–428 (1999).
Hoshino, T. et al. Suppression of Alzheimer’s disease-related phenotypes by expression of heat shock protein 70 in mice. J. Neurosci. 31, 5225–5234 (2011).
Cummings, C. J. et al. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 10, 1511–1518 (2001).
Labbadia, J. et al. Suppression of protein aggregation by chaperone modification of high molecular weight complexes. Brain 135, 1180–1196 (2012).
Sittler, A. et al. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum. Mol. Genet. 10, 1307–1315 (2001). This paper shows that pharmacological induction of the stress response can prevent aggregation of a disease protein.
Nagy, M., Fenton, W. A., Li, D., Furtak, K. & Horwich, A. L. Extended survival of misfolded G85R SOD1-linked ALS mice by transgenic expression of chaperone Hsp110. Proc. Natl Acad. Sci. USA 113, 5424–5428 (2016).
Calamini, B. et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8, 185–196 (2011).
Sontag, E. M. et al. Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc. Natl Acad. Sci. USA 110, 3077–3082 (2013).
Das, I. et al. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348, 239–242 (2015).
Menzies, F. M. et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034 (2017).
Lee, B. H. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 (2010).
Rousseau, A. & Bertolotti, A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536, 184–189 (2016). This paper describes an evolutionarily conserved signalling pathway that controls proteasome homeostasis.
Mendillo, M. L. et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562 (2012).
Joshi, S. et al. Adapting to stress — chaperome networks in cancer. Nat. Rev. Cancer 18, 562–575 (2018).
Calderwood, S. K. & Neckers, L. Hsp90 in cancer: transcriptional roles in the nucleus. Adv. Cancer Res. 129, 89–106 (2016).
Joazeiro, C. A. P. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol. 33, 343–368 (2017).
Bengtson, M. H. & Joazeiro, C. A. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010). This paper presents identification of listerin (Ltn1) as the E3 ligase crucial for the proteasomal degradation of failed nascent polypeptide chains on ribosomes.
Brandman, O. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012).
Shen, P. S. et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 (2015).
Kostova, K. K. et al. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357, 414–417 (2017).
Defenouillere, Q. & Fromont-Racine, M. The ribosome-bound quality control complex: from aberrant peptide clearance to proteostasis maintenance. Curr. Genet. 63, 997–1005 (2017).
Izawa, T., Park, S. H., Zhao, L., Hartl, F. U. & Neupert, W. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell 171, 890–903 (2017).
Nielson, J. R. et al. Sterol oxidation mediates stress-responsive Vms1 translocation to mitochondria. Mol. Cell 68, 673–685 (2017).
Verma, R. et al. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature 557, 446–451 (2018).
Zurita Rendon, O. et al. Vms1p is a release factor for the ribosome-associated quality control complex. Nat. Commun. 9, 2197 (2018).
Feng, J., Bussiere, F. & Hekimi, S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644 (2001).
Dillin, A. et al. Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401 (2002).
Durieux, J., Wolff, S. & Dillin, A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 (2011).
Tian, Y. et al. Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt). Cell 165, 1197–1208 (2016).
Merkwirth, C. et al. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell 165, 1209–1223 (2016).
Labbadia, J. et al. Mitochondrial stress restores the heat shock response and prevents proteostasis collapse during aging. Cell Rep. 21, 1481–1494 (2017).
Acknowledgements
The authors thank D. Balchin, D. Broch-Trentini, G. Jayaraj and C. Klaips for critically reading the manuscript. Work in the authors’ laboratory is supported by the European Commission under FP7 GA ERC-2012-SyG_318987–ToPAG and the Deutsche Forschungsgemeinschaft (German Research Foundation) within the framework of the Munich Cluster for Systems Neurology.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
F.U.H. holds stock options in, receives consulting fees from and is the chair of the scientific advisory board of Proteostasis Therapeutics, Inc. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Intrinsically disordered regions
-
Regions of a protein that lack stable, well-defined tertiary structure; often functionally relevant in interactions with partner proteins.
- Tail-anchored proteins
-
Membrane proteins that are post-translationally inserted into the membrane. They can contain a transmembrane sequence near the carboxy terminus.
- E3 ubiquitin ligase
-
An enzyme that mediates the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to a protein substrate.
- E2 ubiquitin-conjugating enzyme
-
An enzyme that catalyses the second step in the enzymatic cascade for the transfer of ubiquitin to protein substrates.
- Reticulocytes
-
Immature red blood cells.
- Co-chaperone
-
A factor that assists or regulates the function of a molecular chaperone; some co-chaperones also have chaperone activity in binding non-native proteins.
- Chaperone-assisted selective autophagy
-
A degradation pathway of chaperone-bound proteins in lysosomes.
- Chaperone-mediated autophagy
-
A chaperone-dependent degradation pathway of soluble cytosolic proteins that involves translocation of the substrate protein across the lysosomal membrane.
- Endosomal microautophagy
-
Degradation of cytosolic proteins by late endosomes and/or multivesicular bodies.
- Amyloid
-
A fibrillar aggregate, composed of polypeptides forming a cross-β structure, that has defined tinctorial (dye-binding) properties.
- Low-complexity domains
-
Sequences of amino acids with little diversity that are often intrinsically unstructured.
- Polyglutamine expansion
-
Pathogenic elongation of a polyglutamine stretch in a protein caused by an increased number of CAG trinucleotide repeats; described in a group of unrelated genes.
- Chaperonin
-
A class of molecular chaperones forming large, double-ring complexes that transiently enclose a substrate protein for folding (examples include HSP60 in mitochondria and TRiC in the eukaryotic cytosol).
- BRICHOS domain
-
A domain found in several proteins associated with dementia, respiratory distress and cancer, including BRI2, chondromodulin I and surfactant protein C. BRICHOS domains have intramolecular chaperone-like activities and inhibit misfolding and aggregation.
- Hormesis
-
An adaptive response of an organism or biological system towards a low dose of a toxic agent or physical conditions (for example, reactive oxygen radicals or thermal stress) that preconditions the organism to tolerate a higher dose of the same toxic agent.
- Critical concentration
-
The concentration up to which a protein remains soluble; exceeding this concentration results in insolubility and aggregation.
- Unfolded protein response
-
(UPR). A cellular stress response pathway that serves to increase the protein-folding capacity of the endoplasmic reticulum or the mitochondria.
- Integrated stress response
-
A conserved signalling pathway that responds to a variety of cellular conditions and attenuates protein translation via phosphorylation of translation initiation factor 2α (eIF2α).
- Transthyretin
-
A tetrameric transport protein that binds to the thyroid hormone thyroxin and retinol-binding protein. Mutant forms dissociate into subunits and aggregate, resulting in transthyretin amyloidosis.
Rights and permissions
About this article
Cite this article
Hipp, M.S., Kasturi, P. & Hartl, F.U. The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 20, 421–435 (2019). https://doi.org/10.1038/s41580-019-0101-y
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-019-0101-y
This article is cited by
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Emerging role of senescent microglia in brain aging-related neurodegenerative diseases
Translational Neurodegeneration (2024)
-
Stress response silencing by an E3 ligase mutated in neurodegeneration
Nature (2024)
-
Chp1 is a dedicated chaperone at the ribosome that safeguards eEF1A biogenesis
Nature Communications (2024)
-
Tracing genetic diversity captures the molecular basis of misfolding disease
Nature Communications (2024)