Abstract
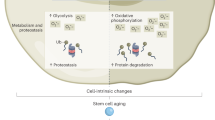
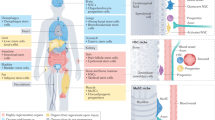
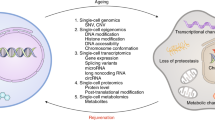
Adult tissue stem cells have a pivotal role in tissue maintenance and regeneration throughout the lifespan of multicellular organisms. Loss of tissue homeostasis during post-reproductive lifespan is caused, at least in part, by a decline in stem cell function and is associated with an increased incidence of diseases. Hallmarks of ageing include the accumulation of molecular damage, failure of quality control systems, metabolic changes and alterations in epigenome stability. In this Review, we discuss recent evidence in support of a novel concept whereby cell-intrinsic damage that accumulates during ageing and cell-extrinsic changes in ageing stem cell niches and the blood result in modifications of the stem cell epigenome. These cumulative epigenetic alterations in stem cells might be the cause of the deregulation of developmental pathways seen during ageing. In turn, they could confer a selective advantage to mutant and epigenetically drifted stem cells with altered self-renewal and functions, which contribute to the development of ageing-associated organ dysfunction and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Martin, N., Beach, D. & Gil, J. Ageing as developmental decay: insights from p16INK4a. Trends Mol. Med. 20, 667–674 (2014).
Henderson, S. T. & Johnson, T. E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11, 1975–1980 (2001).
Blagosklonny, M. V. Aging is not programmed. Cell Cycle 12, 3736–3742 (2013).
Behrens, A., van Deursen, J. M., Rudolph, K. L. & Schumacher, B. Impact of genomic damage and ageing on stem cell function. Nat. Cell Biol. 16, 201–207 (2014).
Mohrin, M. et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185 (2010).
Sotiropoulou, P. A. et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat. Cell Biol. 12, 572–582 (2010).
Goloubinoff, P., Sassi, A. S., Fauvet, B., Barducci, A. & De Los Rios, P. Chaperones convert the energy from ATP into the nonequilibrium stabilization of native proteins. Nat. Chem. Biol. https://doi.org/10.1038/s41589-018-0013-8 (2018).
Peth, A., Nathan, J. A. & Goldberg, A. L. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J. Biol. Chem. 288, 29215–29222 (2013).
Ho, T. T. et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017). This study reports that subpopulations of HSCs exhibit defects in autophagy during ageing that are associated with impairments in stem cell function.
Franceschi, C. et al. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 69 (Suppl. 1), S4–S9 (2014).
Thevaranjan, N. et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 21, 455–466.e4 (2017).
Baylin, S. B. The cancer epigenome: its origins, contributions to tumorigenesis, and translational implications. Proc. Am. Thorac Soc. 9, 64–65 (2012).
Simsek, T. et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010).
Takubo, K. et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61 (2013).
Rodríguez-Colman, M. J. et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543, 424–427 (2017).
Park, C. B. & Larsson, N. G. Mitochondrial DNA mutations in disease and aging. J. Cell Biol. 193, 809–818 (2011).
Garcia-Prat, L. et al. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016).
Ito, K. et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 12, 446–451 (2006).
Owusu-Ansah, E. & Banerjee, U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461, 537–541 (2009).
Mantel, C. R. et al. Enhancing hematopoietic stem cell transplantation efficacy by mitigating oxygen shock. Cell 161, 1553–1565 (2015).
Guo, L., Karpac, J., Tran, S. L. & Jasper, H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 156, 109–122 (2014).
Biteau, B. et al. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 6, e1001159 (2010).
Biteau, B., Hochmuth, C. E. & Jasper, H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 3, 442–455 (2008).
Zhang, H. et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 (2016). This study shows that treatment with NAD + precursor nicotinamide riboside improves stem cell function and prevents their senescence in ageing by enhancing mitochondrial quality.
Ryall, J. G. et al. The NAD+-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16, 171–183 (2015).
Cuervo, A. M. Autophagy and aging: keeping that old broom working. Trends Genet. 24, 604–612 (2008).
Geiger, H., de Haan, G. & Florian, M. C. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 13, 376–389 (2013).
Shpilka, T. & Haynes, C. M. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell. Biol. 19, 109–120 (2017).
Douglas, P. M. & Dillin, A. Protein homeostasis and aging in neurodegeneration. J. Cell Biol. 190, 719–729 (2010).
Lapierre, L. R., Kumsta, C., Sandri, M., Ballabio, A. & Hansen, M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 11, 867–880 (2015).
Merkwirth, C. et al. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell 165, 1209–1223 (2016).
Mohrin, M. et al. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377 (2015).
Solanas, G. et al. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 170, 678–692.e20 (2017).
Cellerino, A. & Ori, A. What have we learned on aging from omics studies? Semin. Cell Dev. Biol. 70, 177–189 (2017).
Buchwalter, A. & Hetzer, M. W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 8, 328 (2017).
Zhang, W. et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348, 1160–1163 (2015).
Signer, R. A. J., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014).
Wang, L., Ryoo, H. D., Qi, Y. & Jasper, H. PERK limits Drosophila lifespan by promoting intestinal stem cell proliferation in response to ER stress. PLoS Genet. 11, e1005220 (2015).
Bradley, E., Bieberich, E., Mivechi, N. F., Tangpisuthipongsa, D. & Wang, G. Regulation of embryonic stem cell pluripotency by heat shock protein 90. Stem Cells 30, 1624–1633 (2012).
Noormohammadi, A. et al. Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat. Commun. 7, 13649 (2016).
Vilchez, D. et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489, 304–308 (2012).
van Galen, P. et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–272 (2014).
Bufalino, M. R., DeVeale, B. & van der Kooy, D. The asymmetric segregation of damaged proteins is stem cell-type dependent. J. Cell Biol. 201, 523–530 (2013).
Katajisto, P. et al. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343 (2015).
Moore, D. L., Pilz, G. A., Arauzo-Bravo, M. J., Barral, Y. & Jessberger, S. A mechanism for the segregation of age in mammalian neural stem cells. Science 349, 1334–1338 (2015).
Erjavec, N., Larsson, L., Grantham, J. & Nystrom, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21, 2410–2421 (2007).
Bernet, J. D. et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 20, 265–271 (2014).
Zou, C. & Mallampalli, R. K. Regulation of histone modifying enzymes by the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 694–702 (2014).
Gambetta, M. C. & Müller, J. O-GlcNAcylation prevents aggregation of the polycomb group repressor polyhomeotic. Dev. Cell 31, 629–639 (2014).
Hammond, C. M., Strømme, C. B., Huang, H., Patel, D. J. & Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 (2017).
Das, C. & Tyler, J. K. Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta 1819, 332–342 (2013).
Feser, J. et al. Elevated histone expression promotes life span extension. Mol. Cell 39, 724–735 (2010).
Sperka, T., Wang, J. & Rudolph, K. L. DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 13, 579–590 (2012).
Burkhalter, M. D., Rudolph, K. L. & Sperka, T. Genome instability of ageing stem cells-Induction and defence mechanisms. Ageing Res. Rev. 23, 29–36 (2015).
Beerman, I., Seita, J., Inlay, M. A., Weissman, I. L. & Rossi, D. J. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15, 37–50 (2014).
Flach, J. et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014). This study shows that DNA damage increases in ageing HSCs as a consequence of increased replication stress owing to imbalances in the expression of components of the DNA replication complex.
Walter, D. et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520, 549–552 (2015). This study shows that entry into the cell cycle from dormancy is a phase of vulnerability in HSCs, leading to an accumulation of DNA damage and stem cell exhaustion.
Rossi, D. J. et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007).
Nijnik, A. et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature 447, 686–690 (2007).
Gorbunova, V. & Seluanov, A. DNA double strand break repair, aging and the chromatin connection. Mutat. Res. Mol. Mech. Mutag. 788, 2–6 (2016).
Kauppinen, T. M., Gan, L. & Swanson, R. A. Poly(ADP-ribose) polymerase-1-induced NAD(+) depletion promotes nuclear factor-κB transcriptional activity by preventing p65 de-acetylation. Biochim. Biophys. Acta 1833, 1985–1991 (2013).
Missios, P. et al. Glucose substitution prolongs maintenance of energy homeostasis and lifespan of telomere dysfunctional mice. Nat. Commun. 5, 4924 (2014).
O’Sullivan, R. J., Kubicek, S., Schreiber, S. L. & Karlseder, J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 17, 1218–1225 (2010).
Liu, L. et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189–204 (2013).
Conboy, I. M. et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433, 760–764 (2005).
Villeda, S. A. et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94 (2011).
Carlson, M. E., Hsu, M. & Conboy, I. M. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature 454, 528–532 (2008).
Sinha, M. et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 344, 649–652 (2014).
Elabd, C. et al. Oxytocin is an age-specific circulating hormone that is necessary for muscle maintenance and regeneration. Nat. Commun. 5, 4082 (2014).
Egerman, M. A. et al. GDF11 increases with age and inhibits skeletal muscle regeneration. Cell Metab. 22, 164–174 (2015).
Castellano, J. M. et al. Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492 (2017).
Gancz, D. & Gilboa, L. Hormonal control of stem cell systems. Annu. Rev. Cell Dev. Biol. 29, 137–162 (2013).
Nakada, D. et al. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature 505, 555–558 (2014).
Kim, J.-H. et al. Sex hormones establish a reserve pool of adult muscle stem cells. Nat. Cell Biol. 18, 930–940 (2016).
Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003).
Ergen, A. V., Boles, N. C. & Goodell, M. A. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood 119, 2500–2509 (2012).
Mossadegh-Keller, N. et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature 497, 239–243 (2013).
Palacios, D. et al. TNF/p38?/Polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7, 455–469 (2010).
Weir, H. J. et al. Dietary restriction and AMPK increase lifespan via mitochondrial network and peroxisome remodeling. Cell Metab. 26, 884–896.e5 (2017).
Smith, P. et al. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. eLife 6, e27014 (2017).
Hahn, O. et al. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 18, 1194 (2017).
Mendelson, A. & Frenette, P. S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 20, 833–846 (2014).
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004).
Morrison, S. J. & Scadden, D. T. The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014).
Blau, H. M., Cosgrove, B. D. & Ho, A. T. V. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 21, 854–862 (2015).
Barker, N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2013).
Kaiko, G. E. et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 165, 1708–1720 (2016).
Chakkalakal, J. V., Jones, K. M., Basson, M. A. & Brack, A. S. The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360 (2012).
Nalapareddy, K. et al. Canonical Wnt signaling ameliorates aging of intestinal stem cells. Cell Rep. 18, 2608–2621 (2017).
Lindemans, C. A. et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015).
Campisi, J. Aging, Cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013).
Herbig, U., Ferreira, M., Condel, L., Carey, D. & Sedivy, J. M. Cellular senescence in aging primates. Science 311, 1257 (2006).
Song, Z., Zhang, J., Ju, Z. & Rudolph, K. L. Telomere dysfunctional environment induces loss of quiescence and inherent impairments of hematopoietic stem cell function. Aging Cell 11, 449–455 (2012).
Guidi, N. et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J. 36, 840–853 (2017).
Lukjanenko, L. et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 22, 897 (2016).
Rozo, M., Li, L. & Fan, C.-M. Targeting β1-integrin signaling enhances regeneration in aged and dystrophic muscle in mice. Nat. Med. 22, 889–896 (2016).
Boyle, M. et al. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell 1, 470–478 (2007).
van Niel, G., D’Angelo, G. & Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018).
Fry, C. S., Kirby, T. J., Kosmac, K., McCarthy, J. J. & Peterson, C. A. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell 20, 56–69 (2017).
Fusco, S. et al. A CREB-Sirt1-Hes1 circuitry mediates neural stem cell response to glucose availability. Cell Rep. 14, 1195–1205 (2016).
Gjorevski, N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016).
Stearns-Reider, K. M. et al. Aging of the skeletal muscle extracellular matrix drives a stem cell fibrogenic conversion. Aging Cell 16, 518–528 (2017).
Crowder, S. W., Leonardo, V., Whittaker, T., Papathanasiou, P. & Stevens, M. M. Material cues as potent regulators of epigenetics and stem cell function. Cell Stem Cell 18, 39–52 (2016).
Buczacki, S. J. A. et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69 (2013).
Rosenberger, G. et al. Statistical control of peptide and protein error rates in large-scale targeted data-independent acquisition analyses. Nat. Methods 14, 921–927 (2017).
Jadhav, U. et al. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell 21, 65–77.e5 (2017).
Basak, O. et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell 20, 177–190.e4 (2017).
Yan, K. S. et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature 545, 238–242 (2017).
Fontana, L. & Partridge, L. Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106–118 (2015).
Tang, D. et al. Dietary restriction improves repopulation but impairs lymphoid differentiation capacity of hematopoietic stem cells in early aging. J. Exp. Med. 213, 535–553 (2016).
Yilmaz, Ö. H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Mair, W., McLeod, C. J., Wang, L. & Jones, D. L. Dietary restriction enhances germline stem cell maintenance. Aging Cell 9, 916–918 (2010).
Cerletti, M., Jang, Y. C., Finley, L. W. S., Haigis, M. C. & Wagers, A. J. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell 10, 515–519 (2012).
Brandhorst, S. et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 22, 86–99 (2015).
Cheng, C.-W. et al. Fasting-mimicking diet promotes Ngn3-driven β-cell regeneration to reverse diabetes. Cell 168, 775–788.e12 (2017).
Regan, J. C. et al. Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. eLife 5, e10956 (2016).
Goldberg, E. L. et al. Lifespan-extending caloric restriction or mTOR inhibition impair adaptive immunity of old mice by distinct mechanisms. Aging Cell 14, 130–138 (2015).
Lazare, S. et al. Lifelong dietary intervention does not affect hematopoietic stem cell function. Exp. Hematol. 53, 26–30 (2017).
Igarashi, M. & Guarente, L. mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell 166, 436–450 (2016).
Zhang, C. et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat. Commun. 4, 2163 (2013).
Petkovich, D. A. et al. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 25, 954–960.e6 (2017).
Öst, A. et al. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159, 1352–1364 (2014).
Cabezas-Wallscheid, N. et al. Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell 169, 807–823.e19 (2017).
Cimmino, L. et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170, 1079–1095.e20 (2017).
Agathocleous, M. et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476 (2017).
D’Aniello, C., Cermola, F., Patriarca, E. J. & Minchiotti, G. Vitamin C in stem cell biology: impact on extracellular matrix homeostasis and epigenetics. Stem Cells Int. 2017, 8936156 (2017).
Booth, L. N. & Brunet, A. The aging epigenome. Mol. Cell 62, 728–744 (2016).
Horvath, S. et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 13, R97 (2012).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
Stubbs, T. M. et al. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 18, 68 (2017).
Weidner, C. et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 15, R24 (2014).
Yuan, T. et al. An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet. 11, e1004996 (2015).
Beerman, I. et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell 12, 413–425 (2013).
Arai, K. et al. Total synthesis of 6-deoxypladienolide D and assessment of splicing inhibitory activity in a mutant SF3B1 cancer cell line. Org. Lett. 16, 5560–5563 (2014).
Fernández, A. F. et al. H3K4me1 marks DNA regions hypomethylated during aging in human stem and differentiated cells. Genome Res. 25, 27–40 (2014).
Cole, J. J. et al. Diverse interventions that extend mouse lifespan suppress shared age-associated epigenetic changes at critical gene regulatory regions. Genome Biol. 18, E503 (2017).
Maegawa, S. et al. Caloric restriction delays age-related methylation drift. Nat. Commun. 8, 539 (2017).
Schwörer, S. et al. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 540, 428–432 (2016). This study shows that epigenetic alteration in response to activation limits the self-renewal and function of aged muscle stem cells by activation of developmental signals.
Sun, D. et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14, 673–688 (2014).
Beerman, I. & Rossi, D. J. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell 16, 613–625 (2015).
Kazakevych, J., Sayols, S., Messner, B., Krienke, C. & Soshnikova, N. Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 45, 5770–5784 (2017).
Kim, K.-M. & Shibata, D. Methylation reveals a niche: stem cell succession in human colon crypts. Oncogene 21, 5441–5449 (2002).
Ro, S. & Rannala, B. Methylation patterns and mathematical models reveal dynamics of stem cell turnover in the human colon. Proc. Natl Acad. Sci. USA 98, 10519–10521 (2001).
Kaaij, L. T. J. et al. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 14, R50 (2013).
Sheaffer, K. L. et al. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 28, 652–664 (2014).
Ciccocioppo, R. et al. Small bowel enterocyte apoptosis and proliferation are increased in the elderly. Gerontology 48, 204–208 (2002).
Corazza, G. R. et al. Proliferating cell nuclear antigen expression is increased in small bowel epithelium in the elderly. Mech. Ageing Dev. 104, 1–9 (1998).
Kim, J. Y., Siegmund, K. D., Tavaré, S. & Shibata, D. Age-related human small intestine methylation: evidence for stem cell niches. BMC Med. 3, 10 (2005).
Camp, J. G. et al. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 24, 1504–1516 (2014).
Hahn, M. A. et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 68, 10280–10289 (2008).
Mugatroyd, C., Wu, Y., Bockmühl, Y. & Spengler, D. The Janus face of DNA methylation in aging. Aging 2, 107–110 (2010).
Takahashi, K. et al. Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J. Biol. Chem. 286, 35755–35762 (2011).
Kirschner, K. et al. Proliferation drives aging-related functional decline in a subpopulation of the hematopoietic stem cell compartment. Cell Rep. 19, 1503–1511 (2017).
Enge, M. et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330.e14 (2017).
Cheung, P. et al. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell https://doi.org/10.1016/j.cell.2018.03.079 (2018).
Alcolea, M. P. et al. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat. Cell Biol. 16, 615–622 (2014).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Goriely, A. & Wilkie, A. O. M. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am. J. Hum. Genet. 90, 175–200 (2012).
Greaves, L. C. et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl Acad. Sci. USA 103, 714–719 (2006).
Hsieh, J. C. F., Van Den Berg, D., Kang, H., Hsieh, C.-L. & Lieber, M. R. Large chromosome deletions, duplications, and gene conversion events accumulate with age in normal human colon crypts. Aging Cell 12, 269–279 (2013).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014).
McKerrell, T. et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 10, 1239–1245 (2015).
Okuchi, Y. et al. Identification of aging-associated gene expression signatures that precede intestinal tumorigenesis. PLoS ONE 11, e0162300 (2016).
Shlush, L. I. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014).
Busque, L. et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 44, 1179–1181 (2012).
Lu, R. et al. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell 30, 92–107 (2016).
Scourzic, L. et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia 30, 1388–1398 (2016).
Tefferi, A. et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 23, 905–911 (2009).
Yang, L. et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell 29, 922–934 (2016).
Challen, G. A. et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44, 23–31 (2011).
Ko, M. et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl Acad. Sci. USA 108, 14566–14571 (2011).
Zhang, X. et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 48, 1014–1023 (2016).
Fuster, J. J. et al. Clonal hematopoiesis associated with Tet2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017).
Jaiswal, S. et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017). References 172 and 173 report that the clonal dominance of HSCs with mutations in epigenome regulators contributes to the development of ageing-associated diseases.
Bahar, R. et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441, 1011–1014 (2006).
Martincorena, I. et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
He, J. & Zhang, Y. Janus kinase 2: an epigenetic ‘writer’ that activates leukemogenic genes. J. Mol. Cell. Biol. 2, 231–233 (2010).
Mian, S. A. et al. Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome. Haematologica 98, 1058–1066 (2013).
Vrba, L., Junk, D. J., Novak, P. & Futscher, B. W. p53 induces distinct epigenetic states at its direct target promoters. BMC Genomics 9, 486 (2008).
Reddington, J. P., Pennings, S. & Meehan, R. R. Non-canonical functions of the DNA methylome in gene regulation. Biochem. J. 451, 13–23 (2013).
De Cecco, M. et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12, 247–256 (2013).
Van Meter, M. et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 5, 5011 (2014).
Neri, F. et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77 (2017).
Kim, J. et al. Blocking promiscuous activation at cryptic promoters directs cell type-specific gene expression. Science 356, 717–721 (2017).
Maunakea, A. K., Chepelev, I., Cui, K. & Zhao, K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 23, 1256–1269 (2013).
Shukla, S. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011).
Yearim, A. et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 10, 1122–1134 (2015).
Irimia, M. & Blencowe, B. J. Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 24, 323–332 (2012).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Logan, C. Y. & Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 (2004).
Clevers, H. Wnt/beta-Catenin signaling in development and disease. Cell 127, 469–480 (2006).
Thisse, B. & Thisse, C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev. Biol. 287, 390–402 (2005).
Kingsley, D. M. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 8, 133–146 (1994).
Keren, A., Tamir, Y. & Bengal, E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 252, 224–230 (2006).
Muñoz-Espín, D. et al. Programmed cell senescence during mammalian embryonic development. Cell 155, 1104–1118 (2013).
Storer, M. et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155, 1119–1130 (2013).
Brack, A. S. et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317, 807–810 (2007).
Yang, L. et al. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc. Natl Acad. Sci. USA 104, 5091–5096 (2007).
Florian, M. C. et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 10, 520–530 (2012).
Famili, F. et al. Discrete roles of canonical and non-canonical Wnt signaling in hematopoiesis and lymphopoiesis. Cell Death Dis. 6, e1981 (2015).
Florian, M. C. et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature 503, 392–396 (2013).
Povinelli, B. J. & Nemeth, M. J. Wnt5a regulates hematopoietic stem cell proliferation and repopulation through the Ryk receptor. Stem Cells 32, 105–115 (2014).
Nemeth, M. J., Topol, L., Anderson, S. M., Yang, Y. & Bodine, D. M. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proc. Natl Acad. Sci. USA 104, 15436–15441 (2007).
Sugimura, R. et al. Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche. Cell 150, 351–365 (2012).
Tao, S. et al. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 34, 624–640 (2015).
Scaffidi, P. & Misteli, T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat. Cell Biol. 10, 452–459 (2008).
Espada, J. et al. Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J. Cell Biol. 181, 27–35 (2008).
Cairney, C. J. et al. A systems biology approach to Down syndrome: Identification of Notch/Wnt dysregulation in a model of stem cells aging. Biochim. Biophys. Acta 1792, 353–363 (2009).
Meena, J. K. et al. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 34, 1371–1384 (2015).
Conboy, I. M., Conboy, M. J., Smythe, G. M. & Rando, T. A. Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577 (2003).
Sousa-Victor, P. et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014).
Duncan, A. W. et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 6, 314–322 (2005).
Janzen, V. et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443, 421–426 (2006).
Chen, K.-Y. et al. A Notch positive feedback in the intestinal stem cell niche is essential for stem cell self-renewal. Mol. Syst. Biol. 13, 927 (2017).
Siudeja, K. et al. Frequent somatic mutation in adult intestinal stem cells drives neoplasia and genetic mosaicism during aging. Cell Stem Cell 17, 663–674 (2015).
van Es, J. H. et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005).
Cosgrove, B. D. et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 20, 255–264 (2014).
de Haan, G. et al. In vitro generation of long-term repopulating hematopoietic stem cells by fibroblast growth factor-1. Dev. Cell 4, 241–251 (2003).
Berent-Maoz, B., Montecino-Rodriguez, E., Signer, R. A. J. & Dorshkind, K. Fibroblast growth factor-7 partially reverses murine thymocyte progenitor aging by repression of Ink4a. Blood 119, 5715–5721 (2012).
Al Alam, D. et al. Fibroblast growth factor 10 alters the balance between goblet and Paneth cells in the adult mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 308, G678–G690 (2015).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011).
Vidrich, A. et al. Fibroblast growth factor receptor-3 regulates Paneth cell lineage allocation and accrual of epithelial stem cells during murine intestinal development. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G168–G178 (2009).
Liu, D.-W., Tsai, S.-M., Lin, B.-F., Jiang, Y.-J. & Wang, W.-P. Fibroblast growth factor receptor 2c signaling is required for intestinal cell differentiation in zebrafish. PLoS ONE 8, e58310 (2013).
Park, J.-S., Kim, Y.-S. & Yoo, M.-A. The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging 1, 637–651 (2009).
Avgustinova, A. & Benitah, S. A. Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell. Biol. 17, 643–658 (2016).
Ocampo, A. et al. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167, 1719–1733.e12 (2016).
Yin, H., Price, F. & Rudnicki, M. A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013).
Nuschke, A., Rodrigues, M., Wells, A. W., Sylakowski, K. & Wells, A. Mesenchymal stem cells/multipotent stromal cells (MSCs) are glycolytic and thus glucose is a limiting factor of in vitro models of MSC starvation. Stem Cell Res. Ther. 7, 179 (2016).
Zhang, J. et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 30, 4860–4873 (2011).
Takubo, K. et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7, 391–402 (2010).
Maryanovich, M. et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat. Commun. 6, 7901 (2015).
Rocheteau, P., Gayraud-Morel, B., Siegl-Cachedenier, I., Blasco, M. A. & Tajbakhsh, S. A. Subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125 (2012).
Moussaieff, A. et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 21, 392–402 (2015).
Imai, S. & Guarente, L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 (2014).
Choudhury, A. R. et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 39, 99–105 (2007).
Baker, D. J. et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
Chang, J. et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016). References 238 and 239 report that the depletion of senescent cells in aged mice improves stem cell function and organ maintenance, leading to increased health span.
Berry, D. C. et al. Cellular aging contributes to failure of cold-induced beige adipocyte formation in old mice and humans. Cell Metab. 25, 166–181 (2016).
Ju, Z. et al. Telomere dysfunction induces environmental alterations limiting hematopoietic stem cell function and engraftment. Nat. Med. 13, 742–747 (2007).
Passos, J. F. et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 6, 347 (2010).
Mosteiro, L. et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 354, aaf4445 (2016).
Demaria, M. et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014).
Schwitalla, S. et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38 (2013).
You, J. S. & Jones, P. A. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20 (2012).
Krivtsov, A. V. & Armstrong, S. A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7, 823–833 (2007).
Yang, L., Rau, R. & Goodell, M. A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 15, 152–165 (2015).
Wallace, D. C. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 51, 440–450 (2010).
Parker, S. J. & Metallo, C. M. Metabolic consequences of oncogenic IDH mutations. Pharmacol. Ther. 152, 54–62 (2015).
Lahtz, C. & Pfeifer, G. P. Epigenetic changes of DNA repair genes in cancer. J. Mol. Cell. Biol. 3, 51–58 (2011).
Nazemalhosseini Mojarad, E., Kuppen, P. J., Aghdaei, H. A. & Zali, M. R. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol. Hepatol. Bed Bench 6, 120–128 (2013).
Ogino, S. et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut 55, 1000–1006 (2006).
Alabert, C. et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–293 (2014).
Stamatoyannopoulos, J. A. et al. Human mutation rate associated with DNA replication timing. Nat. Genet. 41, 393–395 (2009).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Acknowledgements
F.N. is supported by the Alexander von Humboldt Foundation and the German Federal Ministry for Education and Research. K.L.R. is supported by a European Research Council Advanced Grant (StemCellGerontoGenes). The Fritz Lipmann Institute is a member of the Leibniz Association and is financially supported by the federal government of Germany and the State of Thuringia.
Author information
Authors and Affiliations
Contributions
M.E., F.N., A.O. and K.L.R. contributed equally to writing, revising, researching and discussing the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Quiescence
-
A reversible state that can describe non-dividing stem cells. Cells are in the G0 phase of the cell cycle, characterized by very low metabolic, transcriptional and proliferative activities.
- Progenitor cells
-
Cells that are constantly generated from stem cells with reduced self-renewal and high proliferative activity; needed for production of cell types for tissue regeneration and homeostasis.
- HIF1–PKD2/PKD4 pathway
-
A signalling pathway that adjusts cellular metabolism to reduced levels of oxygen (hypoxia), by suppressing the entry of glycolytic metabolites into mitochondria.
- p38
-
A MAPK that regulates expression of innate immune molecules and stress effectors in response to cell-extrinsic and cell-intrinsic stimuli.
- FOXO
-
A subgroup of the forkhead family of transcription factors regulated by the insulin–PI3K–AKT pathway and involved in induction of stress response and survival genes.
- Myeloid bias
-
Myeloid bias designates preferential differentiation of haematopoietic stem cells into myeloid cells, such as monocytes, macrophages, neutrophils, platelets and dendritic cells.
- Co-chaperone
-
A protein that regulates the activity of chaperones, such as heat shock protein 70 (HSP70) and HSP90, during protein folding.
- Polycomb group protein
-
A chromatin remodeller implicated in the timely silencing of developmental genes.
- Nucleotide excision repair
-
(NER). A DNA repair pathway responsible for removal and repair of DNA helix-distorting lesions, such as cyclobutane pyrimidine dimers and pyrimidine (6–4) pyrimidone photoproducts.
- Non-homologous end joining
-
(NHEJ). A DNA repair pathway that repairs double-strand DNA breaks by directly ligating the broken ends without the use of a homologous template (a rapid but error-prone process).
- Heterochronic parabiosis
-
An experimental procedure in which two animals of different age, for example, one young and one old, are surgically connected so that their blood circulations are shared.
- Gut microbiome
-
The ensemble of commensal bacterial strains that inhabit the digestive tract of an organism.
- Exosomes
-
Vesicles released by cells containing RNAs and proteins, which have several functions, including cell-cell communication.
- Fibrosis
-
The excessive deposition of extracellular matrix proteins caused by aberrant tissue repair and chronic inflammation.
- Reserve stem cells
-
Pools of cells already committed to differentiation, generally quiescent, that upon injury can de-differentiate and revert to a stem-like state.
- Epimutations
-
Alterations of a specific chromatin modification in a specific genomic locus.
- Retrotransposon
-
A class of transposable DNA sequences using RNA as a mobile intermediate.
- Spurious transcription
-
An RNA transcription process starting from an intragenic or intergenic region.
- Progeroid mice
-
Mice carrying mutations in genes that are mutated in human progeroid syndromes that are characterized by segmental premature ageing (affecting specific organs).
- Aneuploidy
-
The irregular deviation of chromosome numbers from the normal situation — not including doubling of chromosome numbers as in tetraploidy.
- Dysbiosis
-
A deviation in the composition of bacterial strains and taxa in the microbiome of an organ or organism characterized by decreases in overall complexity and overgrowth of certain species.
- Lateral inhibition
-
A mechanism by which cells of one type enforce a difference in cell fate and/or cell identity in neighbouring cells.
- Thymic involution
-
A reduction in the thymus mass and in the output of naive T lymphocytes.
Rights and permissions
About this article
Cite this article
Ermolaeva, M., Neri, F., Ori, A. et al. Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol Cell Biol 19, 594–610 (2018). https://doi.org/10.1038/s41580-018-0020-3
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-018-0020-3
This article is cited by
-
Age-related noncanonical TRMT6–TRMT61A signaling impairs hematopoietic stem cells
Nature Aging (2024)
-
Tissue mosaicism following stem cell aging: blood as an exemplar
Nature Aging (2024)
-
Kdm6a-CNN1 axis orchestrates epigenetic control of trauma-induced spinal cord microvascular endothelial cell senescence to balance neuroinflammation for improved neurological repair
Bone Research (2024)
-
Blockade of TGF-β signalling alleviates human adipose stem cell senescence induced by native ECM in obesity visceral white adipose tissue
Stem Cell Research & Therapy (2023)
-
IFNγ-Stat1 axis drives aging-associated loss of intestinal tissue homeostasis and regeneration
Nature Communications (2023)