Abstract
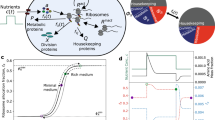
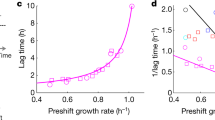
Networks of molecular regulators are often the primary objects of focus in the study of gene regulation, with the machinery of protein synthesis tacitly relegated to the background. Shifting focus to the constraints imposed by the allocation of protein synthesis flux reveals surprising ways in which the actions of molecular regulators are shaped by physiological demands. Using carbon catabolite repression as a case study, we describe how physiological constraints are sensed through metabolic fluxes and how flux-controlled regulation gives rise to simple empirical relations between protein levels and the rate of cell growth.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Neidhardt, F. C., Ingraham, J. L. & Schaechter, M. Physiology of the Bacterial Cell: A Molecular Approach (Sinauer Associates, 1990).
Bervoets, I. & Charlier, D. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol. Rev. 43, 304–339 (2019).
Dorman, C. J. Structure and Function of the Bacterial Genome (Wiley-Blackwell, 2020).
Henkin, T. M. & Peters, J. E. Snyder & Champness Molecular Genetics of Bacteria. 5 edn (ASM Press, 2020).
Phillips, R. The Molecular Switch: Signaling and Allostery (Princeton University Press, 2020).
van den Berg, J., Boersma, A. J. & Poolman, B. Microorganisms maintain crowding homeostasis. Nat. Rev. Microbiol. 15, 309–318 (2017).
Zhang, G. et al. Global and local depletion of ternary complex limits translational elongation. Nucleic Acids Res. 38, 4778–4787 (2010).
Klumpp, S., Scott, M., Pedersen, S. & Hwa, T. Molecular crowding limits translation and cell growth. Proc. Natl Acad. Sci. USA 110, 16754–16759 (2013).
Dai, X. et al. Slowdown of translational elongation in Escherichia coli under hyperosmotic stress. mBio https://doi.org/10.1128/mBio.02375-17 (2018).
Woldringh, C. L., Binnerts, J. S. & Mans, A. Variation in Escherichia coli buoyant density measured in Percoll gradients. J. Bacteriol. 148, 58–63 (1981).
Kubitschek, H. E. Buoyant density variation during the cell cycle in microorganisms. CRC Crit. Rev. Microbiol. 14, 73–97 (1987).
Basan, M. et al. Inflating bacterial cells by increased protein synthesis. Mol. Syst. Biol. 11, 836 (2015).
Oldewurtel, E. R., Kitahara, Y. & van Teeffelen, S. Robust surface-to-mass coupling and turgor-dependent cell width determine bacterial dry-mass density. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2021416118 (2021).
Cayley, S., Lewis, B. A., Guttman, H. J. & Record, M. T. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity: implications for protein-DNA interactions in vivo. J. Mol. Biol. 222, 281–300 (1991).
Milo, R. What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays 35, 1050–1055 (2013).
Balakrishnan, R. et al. Principles of gene regulation quantitatively connect DNA to RNA and proteins in bacteria. bioRxiv https://doi.org/10.1101/2021.05.24.445329 (2021).
Bremer, H. & Dennis, P. P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal https://doi.org/10.1128/ecosal.5.2.3 (2008).
Schmidt, A. et al. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110 (2016).
Mori, M. et al. From coarse to fine: the absolute Escherichia coli proteome under diverse growth conditions. Mol. Syst. Biol. 17, e9536 (2021).
Dai, X. et al. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2, 16231 (2016).
Jun, S., Si, F. W., Pugatch, R. & Scott, M. Fundamental principles in bacterial physiology-history, recent progress, and the future with focus on cell size control: a review. Rep. Prog. Phys. 81, 80 (2018).
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010).
Neidhardt, F. C. & Magasanik, B. Studies on the role of ribonucleic acid in the growth of bacteria. Biochim. Biophys. Acta 42, 99–116 (1960).
You, C. et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500, 301–306 (2013).
Maaloe, O. in Gene Expression Biological Regulation and Development (ed Goldberger, R. F.) 487–542 (Plenum Press, 1979).
Klumpp, S., Zhang, Z. & Hwa, T. Growth rate-dependent global effects on gene expression in bacteria. Cell 139, 1366–1375 (2009).
Hui, S. et al. Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol. Syst. Biol. 11, 784 (2015).
Schaechter, M., Maaloe, O. & Kjeldgaard, N. O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol. 19, 592–606 (1958).
Mairet, F., Gouze, J. L. & de Jong, H. Optimal proteome allocation and the temperature dependence of microbial growth laws. NPJ Syst. Biol. Appl. 7, 14 (2021).
Kaspy, I. et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 4, 3001 (2013).
Chubukov, V., Gerosa, L., Kochanowski, K. & Sauer, U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 12, 327–340 (2014).
Magasanik, B. Catabolite repression. Cold Spring Harb. Symposia Quant. Biol. 26, 249–256 (1961).
Deutscher, J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11, 87–93 (2008).
Gorke, B. & Stulke, J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 (2008).
Epps, H. M. & Gale, E. F. The influence of the presence of glucose during growth on the enzymic activities of Escherichia coli: comparison of the effect with that produced by fermentation acids. Biochem. J. 36, 619–623 (1942).
Ullmann, A. & Monod, J. Cyclic AMP as an antagonist of catabolite repression in Escherichia coli. FEBS Lett. 2, 57–60 (1968).
Perlman, R. & Pastan, I. Cyclic 3’5-AMP: stimulation of beta-galactosidase and tryptophanase induction in E. coli. Biochem. Biophys. Res. Commun. 30, 656–664 (1968).
Zubay, G., Schwartz, D. & Beckwith, J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl Acad. Sci. USA 66, 104–110 (1970).
Saier, M. H. Jr, Feucht, B. U. & Hofstadter, L. J. Regulation of carbohydrate uptake and adenylate cyclase activity mediated by the enzymes II of the phosphoenolpyruvate: sugar phosphotransferase system in Escherichia coli. J. Biol. Chem. 251, 883–892 (1976).
Kolb, A., Busby, S., Buc, H., Garges, S. & Adhya, S. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62, 749–795 (1993).
Postma, P. W., Lengeler, J. W. & Jacobson, G. R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594 (1993).
Saier, M. H. Jr. Regulatory interactions involving the proteins of the phosphotransferase system in enteric bacteria. J. Cell. Biochem. 51, 62–68 (1993).
Deutscher, J., Francke, C. & Postma, P. W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 (2006).
Epstein, W., Rothman-Denes, L. B. & Hesse, J. Adenosine 3’:5’-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc. Natl Acad. Sci. USA 72, 2300–2304 (1975).
Hogema, B. M. et al. Catabolite repression by glucose 6-phosphate, gluconate and lactose in Escherichia coli. Mol. Microbiol. 24, 857–867 (1997).
Bettenbrock, K. et al. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. 189, 6891–6900 (2007).
McFall, E. & Magasanik, B. Effects of thymine and of phosphate deprivation on enzyme synthesis in Escherichia coli. Biochim. Biophys. Acta 55, 900–908 (1962).
Clark, D. J. & Marr, A. G. Studies on the repression of beta-galactosidase in Escherichia coli. Biochim. Biophys. Acta 92, 85–94 (1964).
Mandelstam, J. The repression of constitutive beta-galactosidase in Escherichia coli by glucose and other carbon sources. Biochem. J. 82, 489–493 (1962).
Magasanik, B. & Neidhardt, F. C. Inhibitory effect of glucose on enzyme formation. Nature 178, 801–802 (1956).
Ullmann, A. Catabolite repression: a story without end. Res. Microbiol. 147, 455–458 (1996).
Wanner, B. L., Kodaira, R. & Neidhardt, F. C. Regulation of lac operon expression: reappraisal of the theory of catabolite repression. J. Bacteriol. 136, 947–954 (1978).
Magasanik, B. & Neidhardt, F. C. The effect of glucose on the induced biosynthesis of bacterial enzymes in the presence and absence of inducing agents. Biochim. Biophys. Acta 21, 324–334 (1956).
Dourado, H., Mori, M., Hwa, T. & Lercher, M. J. On the optimality of the enzyme-substrate relationship in bacteria. PLoS Biol. 19, e3001416 (2021).
Mori, M., Hwa, T., Martin, O. C., De Martino, A. & Marinari, E. Constrained allocation flux balance analysis. PLoS Comput. Biol. 12, e1004913 (2016).
Scott, M., Klumpp, S., Mateescu, E. M. & Hwa, T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 10, 747 (2014).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015).
Paul, B. J., Ross, W., Gaal, T. & Gourse, R. L. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38, 749–770 (2004).
Wu, C. et al. Cellular perception of growth rate and the mechanistic origin of bacterial growth laws. Proc. Natl Acad. Sci. USA 119, e2201585119 (2022).
Umbarger, H. E. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 47, 532–606 (1978).
Reitzer, L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57, 155–176 (2003).
Huergo, L. F. & Dixon, R. The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol. Mol. Biol. Rev. 79, 419–435 (2015).
Kochanowski, K. et al. Global coordination of metabolic pathways in Escherichia coli by active and passive regulation. Mol. Syst. Biol. 17, e10064 (2021).
Varma, A. & Palsson, B. O. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60, 3724–3731 (1994).
Kochanowski, K. et al. Functioning of a metabolic flux sensor in Escherichia coli. Proc. Natl Acad. Sci. USA 110, 1130–1135 (2013).
Bennett, B. D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009).
Hu, X. P., Dourado, H., Schubert, P. & Lercher, M. J. The protein translation machinery is expressed for maximal efficiency in Escherichia coli. Nat. Commun. 11, 5260 (2020).
Marr, A. G. Growth rate of Escherichia coli. Microbiol. Rev. 55, 316–333 (1991).
Li, S. H. et al. Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 3, 939–947 (2018).
Prossliner, T., Gerdes, K., Sorensen, M. A. & Winther, K. S. Hibernation factors directly block ribonucleases from entering the ribosome in response to starvation. Nucleic Acids Res. 49, 2226–2239 (2021).
Monod, J. in Selected Papers in Molecular Biology by Jacques Monod (eds Lwoff, A. & Ullmann, A.) (Academic Press, 1978).
Hermsen, R., Okano, H., You, C., Werner, N. & Hwa, T. A growth-rate composition formula for the growth of E.coli on co-utilized carbon substrates. Mol. Syst. Biol. 11, 801 (2015).
Okano, H., Hermsen, R., Kochanowski, K. & Hwa, T. Regulation of hierarchical and simultaneous carbon-substrate utilization by flux sensors in Esherichia coli. Nat. Microbiol. 5, 206–215 (2020).
Wang, X., Xia, K., Yang, X. & Tang, C. Growth strategy of microbes on mixed carbon sources. Nat. Commun. 10, 1279 (2019).
de Groot, D. H., Hulshof, J., Teusink, B., Bruggeman, F. J. & Planque, R. Elementary Growth Modes provide a molecular description of cellular self-fabrication. PLoS Comput. Biol. 16, e1007559 (2020).
Okano, H., Hermsen, R. & Hwa, T. Hierarchical and simultaneous utilization of carbon substrates: mechanistic insights, physiological roles, and ecological consequences. Curr. Opin. Microbiol. 63, 172–178 (2021).
Hwa, T. in The Physics of Living Matter: Space, Time and Information (eds Gross, D., Sevrin, A. & Shraiman, B.) 87–98 (World Scientific Publishing Co., 2020).
Erickson, D. W. et al. A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature 551, 119–123 (2017).
Yuan, J., Fowler, W. U., Kimball, E., Lu, W. & Rabinowitz, J. D. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat. Chem. Biol. 2, 529–530 (2006).
Basan, M. et al. A universal trade-off between growth and lag in fluctuating environments. Nature https://doi.org/10.1038/s41586-020-2505-4 (2020).
Balakrishnan, R., de Silva, R. T., Hwa, T. & Cremer, J. Suboptimal resource allocation in changing environments constrains response and growth in bacteria. Mol. Syst. Biol. 17, e10597 (2021).
Lengeler, J. W. in Regulation of Gene Expression in Escherichia coli (eds Lin, E. C. C. & Lynch, A. S.) Ch. 11, 231–254 (Chapman and Hall, 1996).
Magasanik, B. in The Lactose Operon (eds Beckwith, J. & Zipser, D.) 189–219 (Cold Spring Harbor Laboratory, 1970).
Pavlov, M. Y. & Ehrenberg, M. Optimal control of gene expression for fast proteome adaptation to environmental change. Proc. Natl Acad. Sci. USA 110, 20527–20532 (2013).
Riley, M., Pardee, A. B., Jacob, F. & Monod, J. On the expression of a structural gene. J. Mol. Biol. 2, 216–225 (1960).
Bren, A. et al. Glucose becomes one of the worst carbon sources for E.coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci. Rep. 6, 24834 (2016).
Towbin, B. D. et al. Optimality and sub-optimality in a bacterial growth law. Nat. Commun. 8, 14123 (2017).
Muller, S., Regensburger, G. & Steuer, R. Enzyme allocation problems in kinetic metabolic networks: optimal solutions are elementary flux modes. J. Theor. Biol. 347, 182–190 (2014).
Bruggeman, F. J., Planque, R., Molenaar, D. & Teusink, B. Searching for principles of microbial physiology. FEMS Microbiol. Rev. https://doi.org/10.1093/femsre/fuaa034 (2020).
Dourado, H. & Lercher, M. J. An analytical theory of balanced cellular growth. Nat. Commun. 11, 1226 (2020).
Potrykus, K., Murphy, H., Philippe, N. & Cashel, M. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575 (2011).
Sanchez-Vazquez, P., Dewey, C. N., Kitten, N., Ross, W. & Gourse, R. L. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl Acad. Sci. USA 116, 8310–8319 (2019).
Hengge-Aronis, R. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4, 341–346 (2002).
Richter, K., Haslbeck, M. & Buchner, J. The heat shock response: life on the verge of death. Mol. Cell 40, 253–266 (2010).
Imlay, J. A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 (2013).
Si, F. et al. Mechanistic origin of cell-size control and homeostasis in bacteria. Curr. Biol. 29, 1760–1770.e7 (2019).
Zheng, H. et al. General quantitative relations linking cell growth and the cell cycle in Escherichia coli. Nat. Microbiol. 5, 995–1001 (2020).
Colin, A., Micali, G., Faure, L., Cosentino Lagomarsino, M. & van Teeffelen, S. Two different cell-cycle processes determine the timing of cell division in Escherichia coli. eLife https://doi.org/10.7554/eLife.67495 (2021).
Cooper, S. On the fiftieth anniversary of the Schaechter, Maaloe, Kjeldgaard experiments: implications for cell-cycle and cell-growth control. Bioessays 30, 1019–1024 (2008).
Gefen, O., Fridman, O., Ronin, I. & Balaban, N. Q. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc. Natl Acad. Sci. USA 111, 556–561 (2014).
Kaplan, Y. et al. Observation of universal ageing dynamics in antibiotic persistence. Nature 600, 290–294 (2021).
Biselli, E., Schink, S. J. & Gerland, U. Slower growth of Escherichia coli leads to longer survival in carbon starvation due to a decrease in the maintenance rate. Mol. Syst. Biol. 16, e9478 (2020).
Krasny, L. & Gourse, R. L. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23, 4473–4483 (2004).
Muller, A. L. et al. An alternative resource allocation strategy in the chemolithoautotrophic archaeon Methanococcus maripaludis. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2025854118 (2021).
Atkinson, G. C., Tenson, T. & Hauryliuk, V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6, e23479 (2011).
Boutte, C. C. & Crosson, S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 21, 174–180 (2013).
Zavrel, T. et al. Quantitative insights into the cyanobacterial cell economy. eLife https://doi.org/10.7554/eLife.42508 (2019).
Costello, A. & Badran, A. H. Synthetic biological circuits within an orthogonal central dogma. Trends Biotechnol. 39, 59–71 (2021).
Kim, J., Darlington, A., Salvador, M., Utrilla, J. & Jimenez, J. I. Trade-offs between gene expression, growth and phenotypic diversity in microbial populations. Curr. Opin. Biotechnol. 62, 29–37 (2020).
Cardinale, S. & Arkin, A. P. Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol. J. 7, 856–866 (2012).
Brophy, J. A. & Voigt, C. A. Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014).
Qian, Y., Huang, H. H., Jimenez, J. I. & Del Vecchio, D. Resource competition shapes the response of genetic circuits. ACS Synth. Biol. 6, 1263–1272 (2017).
Weisse, A. Y., Oyarzun, D. A., Danos, V. & Swain, P. S. Mechanistic links between cellular trade-offs, gene expression, and growth. Proc. Natl Acad. Sci. USA 112, E1038–E1047 (2015).
Braniff, N., Scott, M. & Ingalls, B. Component characterization in a growth-dependent physiological context: optimal experimental design. Processes 7, 23 (2019).
Ronne, H. Glucose repression in fungi. Trends Genet. 11, 12–17 (1995).
Compagno, C., Dashko, S. & Piskur, J. in Molecular Mechanisms in Yeast Carbon Metabolism (eds Compagno, C. & Piskur, J.) 1–21 (Springer, 2014).
Kafri, M., Metzl-Raz, E., Jonas, F. & Barkai, N. Rethinking cell growth models. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fow081 (2016).
Boer, V. M., Crutchfield, C. A., Bradley, P. H., Botstein, D. & Rabinowitz, J. D. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol. Biol. Cell 21, 198–211 (2010).
Metzl-Raz, E. et al. Principles of cellular resource allocation revealed by condition-dependent proteome profiling. eLife https://doi.org/10.7554/eLife.28034 (2017).
Hackett, S. R. et al. Systems-level analysis of mechanisms regulating yeast metabolic flux. Science https://doi.org/10.1126/science.aaf2786 (2016).
Brown, C. M. & Rose, A. H. Effects of temperature on composition and cell volume of Candida utilis. J. Bacteriol. 97, 261–270 (1969).
Alberghina, F. A., Sturani, E. & Gohlke, J. R. Levels and rates of synthesis of ribosomal ribonucleic acid, transfer ribonucleic acid, and protein in Neurospora crassa in different steady states of growth. J. Biol. Chem. 250, 4381–4388 (1975).
Kochanowski, K. et al. Systematic alteration of in vitro metabolic environments reveals empirical growth relationships in cancer cell phenotypes. Cell Rep. 34, 108647 (2021).
Hecht, K. A., O’Donnell, A. F. & Brodsky, J. L. The proteolytic landscape of the yeast vacuole. Cell Logist. 4, e28023 (2014).
Tyo, K. E., Liu, Z., Magnusson, Y., Petranovic, D. & Nielsen, J. Impact of protein uptake and degradation on recombinant protein secretion in yeast. Appl. Microbiol. Biotechnol. 98, 7149–7159 (2014).
Armstrong, J. Yeast vacuoles: more than a model lysosome. Trends Cell Biol. 20, 580–585 (2010).
Hays, S. G., Yan, L. L. W., Silver, P. A. & Ducat, D. C. Synthetic photosynthetic consortia define interactions leading to robustness and photoproduction. J. Biol. Eng. 11, 4 (2017).
Chuang, J. S., Frentz, Z. & Leibler, S. Homeorhesis and ecological succession quantified in synthetic microbial ecosystems. Proc. Natl Acad. Sci. USA 116, 14852–14861 (2019).
Amarnath, K. et al. Stress-induced cross-feeding of internal metabolites provides a dynamic mechanism of microbial cooperation. bioRxiv https://doi.org/10.1101/2021.06.24.449802 (2021).
Pinheiro, F., Warsi, O., Andersson, D. I. & Lassig, M. Metabolic fitness landscapes predict the evolution of antibiotic resistance. Nat. Ecol. Evol. https://doi.org/10.1038/s41559-021-01397-0 (2021).
Reitzer, L. Biosynthesis of glutamate, aspartate, asparagine, L-alanine, and D-alanine. EcoSal https://doi.org/10.1128/ecosalplus.3.6.1.3 (2004).
Goldman, E. & Jakubowski, H. Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol. Microbiol. 4, 2035–2040 (1990).
Kotte, O., Zaugg, J. B. & Heinemann, M. Bacterial adaptation through distributed sensing of metabolic fluxes. Mol. Syst. Biol. 6, 355 (2010).
Winkler, M. E. & Ramos-Montanez, S. Biosynthesis of histidine. EcoSal https://doi.org/10.1128/ecosalplus.3.6.1.9 (2009).
Irving, S. E., Choudhury, N. R. & Corrigan, R. M. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271 (2021).
Magnusson, L. U., Farewell, A. & Nystrom, T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13, 236–242 (2005).
Acknowledgements
This Review was shaped by extended discussion with numerous colleagues and collaborators over the years. It grew from early discussions with Eduard Mateescu and Stefan Klumpp, and with Hans Bremer, Lazlo Csonka, Antoine Danchin, Patrick Dennis, Peter Geiduschek, Sydney Kustu, Bill Loomis, Elio Schaechter, and Dalai Yan. Many insightful ideas came from colleagues whose proteomic data underlies the pie charts shown in the figures: Ruedi Aebersold, Gene-wei Li, Christina Ludwig, and especially Vadim Patsalo, Josh Silverman and Jamie Williamson. Our current understanding of proteome allocation constraints would not have been possible without the input of Rosalind Allen, Frank Bruggeman, Mans Ehrenberg, Suckjoon Jun, Meriem el Karoui, Karl Kochanowski, Martin Lercher, Fernanda Pinheiro, Uwe Sauer, Bas Teusink, Yiping Wang, and current and former members of the Hwa laboratory, especially Rohan Balakrishnan, Markus Basan, David Erickson, Tony Hui, Matteo Mori, Hiroyuki Okano, Severin Schink, Chenhao Wu, Conghui You and Zhongge Zhang. Support for the Hwa laboratory has been provided by the NIH (R01GM095903, R01GM109069), the NSF (PHY105873, MCB 1818384) and the Simons Foundation (330378). M.S. was supported by NSERC (2016-03658).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Kerwyn Casey Huang, who co-reviewed with Handuo Shi; Joshua Rabinowitz; and Uwe Sauer for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Anabolic enzymes
-
Enzymes responsible for biosynthesis, including amino acid and nucleotide synthesis.
- Carbon catabolic proteins
-
Proteins responsible for the transport and breakdown of extracellular carbon sources. Operationally, these are genes regulated by cAMP–Crp.
- C-line
-
The negative correlation between catabolic enzyme expression and growth rate in minimal media when growth rate is modulated by carbon source.
- Diauxic growth
-
Multiple stages of exponential growth as carbon sources are preferentially utilized. The time to switch between carbon sources can take several hours.
- Nutrient quality
-
The exponential growth rate can be modulated by changing the carbon source or nitrogen source, or enriching the medium with amino acids and nucleotides.
- Per-total protein-mass abundance
-
The concentration constraint (biomass/cell water volume) allows a direct conversion between the concentration ((amount of a particular molecule)/(cell water volume)) and its abundance relative to total protein mass ((amount of a particular molecule)/(total protein mass)) assuming that the biomass contains a fixed fraction of protein.
- Protein density
-
The buoyant density ((biomass and water mass)/cell volume) is independent of the growth rate under isotonic conditions. A constant density constraint (biomass/cell volume) therefore implies a constant concentration constraint (biomass/cell water volume). The cellular volume has strong growth-rate dependence; consequently, we do not use protein-number-per-cell as an abundance measure in this Review.
- Protein mass fraction
-
The total number of a particular protein is proportional to its protein mass. Using the per-total protein-mass abundance defined above, the protein mass fraction (protein mass of a particular protein/total protein mass) is therefore a direct measure of concentration.
- Proteome
-
The set of all expressed proteins in a given growth condition.
- Proteome sectors
-
Subsets of the proteome that exhibit similar growth-rate dependence under various growth conditions.
- R-line
-
The positive correlation between the abundance of protein-synthetic machinery (chiefly ribosomes) and growth rate when growth rate is modulated by nutrient quality.
- Unregulated protein
-
Protein expression not subject to any transcriptional or post-transcriptional regulation.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Scott, M., Hwa, T. Shaping bacterial gene expression by physiological and proteome allocation constraints. Nat Rev Microbiol 21, 327–342 (2023). https://doi.org/10.1038/s41579-022-00818-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00818-6
This article is cited by
-
Deep mutational scanning reveals the molecular determinants of RNA polymerase-mediated adaptation and tradeoffs
Nature Communications (2023)
-
Functional decomposition of metabolism allows a system-level quantification of fluxes and protein allocation towards specific metabolic functions
Nature Communications (2023)
-
Enzyme expression kinetics by Escherichia coli during transition from rich to minimal media depends on proteome reserves
Nature Microbiology (2023)