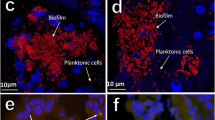
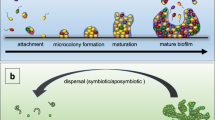
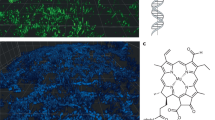
Abstract
Bacterial biofilms are often defined as communities of surface-attached bacteria and are typically depicted with a classic mushroom-shaped structure characteristic of Pseudomonas aeruginosa. However, it has become evident that this is not how all biofilms develop, especially in vivo, in clinical and industrial settings, and in the environment, where biofilms often are observed as non-surface-attached aggregates. In this Review, we describe the origin of the current five-step biofilm development model and why it fails to capture many aspects of bacterial biofilm physiology. We aim to present a simplistic developmental model for biofilm formation that is flexible enough to include all the diverse scenarios and microenvironments where biofilms are formed. With this new expanded, inclusive model, we hereby introduce a common platform for developing an understanding of biofilms and anti-biofilm strategies that can be tailored to the microenvironment under investigation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Costerton, J. W., Geesey, G. G. & Cheng, K. J. How bacteria stick. Sci. Am. 238, 86–95 (1978).
McCoy, W. F., Bryers, J. D., Robbins, J. & Costerton, J. W. Observations of fouling biofilm formation. Can. J. Microbiol. 27, 910–917 (1981).
Flemming, H.-C. et al. Who put the film in biofilm? The migration of a term from wastewater engineering to medicine and beyond. NPJ Biofilms Microbiomes 7, 10 (2021).
Lebeaux, D., Chauhan, A., Rendueles, O. & Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2013, 288–356 (2013).
Hoiby, N. et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 21 (Suppl. 1), S1–S25 (2015).
Kolpen, M. et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax https://doi.org/10.1136/thoraxjnl-2021-217576 (2022).
Raghupathi, P. K. et al. Synergistic interactions within a multispecies biofilm enhance individual species protection against grazing by a pelagic protozoan. Front. Microbiol. https://doi.org/10.3389/fmicb.2017.02649 (2018).
Jass, J., Roberts, S. K. & Lappin-Scott, H. M. in Enzymes in the Environment. Activity, Ecology and Applications 307–326 (Marcel Dekker Inc., 2002).
Tkacz, A. & Poole, P. The plant microbiome: the dark and dirty secrets of plant growth. Plants People Planet 3, 124–129 (2021).
Annous, B. A., Solomon, E. B., Cooke, P. H. & Burke, A. Biofilm formation by Salmonella spp. on Cantaloupe melons. J. Food Saf. 25, 276–287 (2005).
Rudrappa, T., Biedrzycki, M. L. & Bais, H. P. Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 64, 153–166 (2008).
Flemming, H. C. & Wuertz, S. Bacteria and archaea on earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260 (2019). Excellent review that quantitatively explores and proves the long-standing notion that biofilm is the predominant form of prokaryotic life.
Costerton, J. W. et al. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464 (1987).
Vishwakarma, V. Impact of environmental biofilms: industrial components and its remediation. J. Basic Microbiol. 60, 198–206 (2020).
Jurelevicius, D. et al. Long-term souring treatment using nitrate and biocides in high-temperature oil reservoirs. Fuel 288, 119731 (2021).
Alhede, M. et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS ONE 6, e27943 (2011).
Vitzilaiou, E., Kuria, A. M., Siegumfeldt, H., Rasmussen, M. A. & Knøchel, S. The impact of bacterial cell aggregation on UV inactivation kinetics. Water Res. 204, 117593 (2021).
Bjarnsholt, T. et al. The importance of understanding the infectious microenvironment. Lancet Infect. Dis. 22, e88–e92 (2022).
Cornforth, D. M., Diggle, F. L., Melvin, J. A., Bomberger, J. M. & Whiteley, M. Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio https://doi.org/10.1128/mBio.03042-19 (2020).
Marrie, T. J., Nelligan, J. & Costerton, J. W. A scanning and transmission electron microscopic study of an infected endocardial pacemaker lead. Circulation 66, 1339–1341 (1982).
Zobell, C. E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 46, 39–56 (1943). Simple observation results in a paradigm-changing idea: bacteria like to live in communities.
Thaarup, I. C. & Bjarnsholt, T. Current in vitro biofilm-infected chronic wound models for developing new treatment possibilities. Adv. Wound Care 10, 91–102 (2021).
Sternberg, C., Bjarnsholt, T. & Shirtliff, M. Methods for dynamic investigations of surface-attached in vitro bacterial and fungal biofilms. Methods Mol. Biol. 1147, 3–22 (2014).
Azeredo, J. et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 43, 313–351 (2017).
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004).
Irie, Y. et al. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 109, 20632–20636 (2012).
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W. & Davies, D. G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 (2002). The original key paper that started the concept of a biofilm life cycle and is revisited in this Review.
Pamp, S. J., Sternberg, C. & Tolker-Nielsen, T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytom. A 75, 90–103 (2009).
Klausen, M. et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48, 1511–1524 (2003).
Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002).
Moormeier, D. E. & Bayles, K. W. Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol. 104, 365–376 (2017).
Vlamakis, H., Chai, Y., Beauregard, P., Losick, R. & Kolter, R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168 (2013).
Hu, Y., Xiao, Y., Liao, K., Leng, Y. & Lu, Q. Development of microalgal biofilm for wastewater remediation: from mechanism to practical application. J. Chem. Technol. Biotechnol. 96, 2993–3008 (2021).
Liu, J., Lu, H., Wu, L., Kerr, P. G. & Wu, Y. Interactions between periphytic biofilms and dissolved organic matter at soil-water interface and the consequent effects on soil phosphorus fraction changes. Sci. Total Environ. 801, 149708 (2021).
Wu, B. C. et al. Human organoid biofilm model for assessing antibiofilm activity of novel agents. NPJ Biofilms Microbiomes 7, 8 (2021).
Zhao, Y., Liu, H., Wang, R. & Wu, C. Interactions between dicyandiamide and periphytic biofilms in paddy soils and subsequent effects on nitrogen cycling. Sci. Total Environ. 718, 137417 (2020).
Petrova, O. E. & Sauer, K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5, e1000668 (2009). Findings reported here demonstrated, for the first time, that the formation of biofilms is coordinated by a genetic pathway that regulates morphological changes of biofilms and stage-specific transitions in a hierarchically ordered manner. Components of the genetic pathways only seemed to have a role under biofilm growth conditions.
Petrova, O. E., Gupta, K., Liao, J., Goodwine, J. S. & Sauer, K. Divide and conquer: the Pseudomonas aeruginosa two-component hybrid SagS enables biofilm formation and recalcitrance of biofilm cells to antimicrobial agents via distinct regulatory circuits. Environ. Microbiol. 19, 2005–2024 (2017).
O’Toole, G. A. & Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461 (1998).
Davey, M. E. & O’Toole, G. A. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867 (2000).
Characklis, W. G. Attached microbial growths-II. Frictional resistance due to microbial slimes. Water Res. 7, 1249–1258 (1973).
Davies, D. G., Charabarty, A. M. & Geesey, G. G. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 59, 1181–1186 (1993).
Davies, D. G. & Geesey, G. G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61, 860–867 (1995).
Colvin, K. M. et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928 (2012).
Bagge, N. et al. Dynamics and spatial distribution of β-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 48, 1168–1174 (2004).
Wood, D. W., Gong, F., Daykin, M. M., Williams, P. & Pierson, L. S. N-acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J. Bacteriol. 179, 7663–7670 (1997).
Gupta, K., Marques, C. N. H., Petrova, O. E. & Sauer, K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J. Bacteriol. 195, 4975–4987 (2013). Findings reported here indicated that biofilm cells gain heightened tolerance to antimicrobial agents in a manner independent of biofilm biomass accumulation (demonstrating mature biofilm architecture).
Wood, S. R. et al. Architecture of intact natural human plaque biofilms studied by confocal laser scanning microscopy. J. Dent. Res. 79, 21–27 (2000).
Davies, D. G. et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 (1998).
Lequette, Y. & Greenberg, E. P. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187, 37–44 (2005).
Espinosa-Urgel, M. Resident parking only: rhamnolipids maintain fluid channels in biofilms. J. Bacteriol. 185, 699–700 (2003).
Kuchma, S. L., Connolly, J. P. & O’Toole, G. A. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187, 1441–1454 (2005).
Petrova, O. E., Schurr, J. R., Schurr, M. J. & Sauer, K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 86, 819–835 (2012).
Sriramulu, D. D., Lünsdorf, H., Lam, J. S. & Römling, U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54, 667–676 (2005).
Purevdorj, B., Costerton, J. W. & Stoodley, P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68, 4457–4464 (2002). Understanding the relationship between biofilm and fluid dynamics is crucial and completely overlooked in traditional microbiological systems. It impacts not only mass transfer rates but also the biofilm architecture, spatial organization and detachment.
Stewart, P. S. & Franklin, M. J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210 (2008).
Serra, D. O. & Hengge, R. Stress responses go three dimensional–the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 16, 1455–1471 (2014).
Williamson, K. S. et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194, 2062–2073 (2012).
Heacock-Kang, Y. et al. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Mol. Microbiol. 106, 976–985 (2017).
Haussler, S. & Fuqua, C. Biofilms 2012: new discoveries and significant wrinkles in a dynamic field. J. Bacteriol. 195, 2947–2958 (2013).
Rumbaugh, K. P. & Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586 (2020).
Petrova, O. E. & Sauer, K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microbiol. 30, 67–78 (2016).
Davies, D. G. in Biofilm Highlights (eds Flemming, H.-C., Wingender, J. & Szewzyk, U.) 1–28 (Springer, 2011).
Steinberg, N. et al. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Sci. Signal. 13, eaaw8905 (2020).
Purevdorj-Gage, B., Costerton, W. J. & Stoodley, P. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151, 1569–1576 (2005).
Valentini, M. & Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555 (2016).
Römling, U., Galperin, M. Y. & Gomelsky, M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 (2013).
Jenal, U., Reinders, A. & Lori, C. Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017).
Purcell, E. B. & Tamayo, R. Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiol. Rev. 40, 753–773 (2016).
Yin, W., Wang, Y., Liu, L. & He, J. Biofilms: the microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 20, 3423 (2019).
Mantzorou, A. & Ververidis, F. Microalgal biofilms: a further step over current microalgal cultivation techniques. Sci. Total Environ. 651, 3187–3201 (2019).
Prieto-Barajas, C. M., Valencia-Cantero, E. & Santoyo, G. Microbial mat ecosystems: structure types, functional diversity, and biotechnological application. Electron. J. Biotechnol. 31, 48–56 (2018).
Hao, Y. et al. Influence of dental prosthesis and restorative materials interface on oral biofilms. Int. J. Mol. Sci. 19, 3157 (2018).
Chang, C.-S. & Kao, C.-Y. Current understanding of the gut microbiota shaping mechanisms. J. Biomed. Sci. 26, 59 (2019).
Pii, Y. et al. Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils https://doi.org/10.1007/s00374-015-0996-1 (2015).
Roth-Schulze, A. J. et al. Functional biogeography and host specificity of bacterial communities associated with the Marine Green Alga Ulva spp. Mol. Ecol. 27, 1952–1965 (2018).
Monds, R. D. & O’Toole, G. A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17, 73–87 (2009).
Dworkin, M. Developmental Biology of the Bacteria (Benjamin/Cummings Pub. Co., 1985).
Brun, Y. V. & Shimkets, L. J. Prokaryotic Development 114 (ASM Press, 2000).
Goodman, A. L. et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754 (2004).
Merritt, J. H., Brothers, K. M., Kuchma, S. L. & O’Toole, G. A. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189, 8154–8164 (2007).
Chambers, J. R. & Sauer, K. Small RNAs and their role in biofilm formation. Trends Microbiol. 21, 39–49 (2013).
Petrova, O. E., Schurr, J. R., Schurr, M. J. & Sauer, K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol. Microbiol 81, 767–783 (2011).
Petrova, O. E. & Sauer, K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 192, 5275–5288 (2010).
Petrova, O. E. & Sauer, K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 193, 6614–6628 (2011).
Lee, K. W. K. et al. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 8, 894–907 (2014).
Barken, K. B. et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343 (2008).
Friedman, L. & Kolter, R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690 (2004).
Fabbri, S. et al. Fluid-driven interfacial instabilities and turbulence in bacterial biofilms. Env. Microbiol. 19, 4417–4431 (2017).
James, G. A., Beaudette, L. & Costerton, J. W. Interspecies bacterial interactions in biofilms. J. Ind. Microbiol. 15, 257–262 (1995).
Murga, R., Stewart, P. S. & Daly, D. Quantitative analysis of biofilm thickness variability. Biotechnol. Bioeng. 45, 503–510 (1995).
Liu, W. et al. Low-abundant species facilitates specific spatial organization that promotes multispecies biofilm formation. Env. Microbiol. 19, 2893–2905 (2017).
Sauer, K., Steczko, J. & Ash, S. R. Effect of a solution containing citrate/methylene blue/parabens on Staphylococcus aureus bacteria and biofilm, and comparison with various heparin solutions. J. Antimicrob. Chemother. 63, 937–945 (2009).
Allegrucci, M. et al. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188, 2325–2335 (2006).
Hall-Stoodley, L. et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296, 202–211 (2006). First paper to directly link biofilms with otitis media using molecular probes specific for three pathogens, which cause otitis media, on human middle ear mucosal epithelial biopsy samples from children with chronic otitis media and not on uninfected biopsy samples.
Hall-Stoodley, L. et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 8, 173 (2008).
Bakaletz, L. O. Bacterial biofilms in the upper airway - evidence for role in pathology and implications for treatment of otitis media. Paediatr. Respir. Rev. 13, 154–159 (2012).
Walker, W. T. et al. Primary ciliary dyskinesia ciliated airway cells show increased susceptibility to Haemophilus influenzae biofilm formation. Eur. Respir. J. https://doi.org/10.1183/13993003.00612-2017 (2017).
Bjarnsholt, T. et al. The in vivo biofilm. Trends Microbiol. 21, 466–474 (2013).
Tuck, B., Watkin, E., Somers, A. & Machuca, L. L. A critical review of marine biofilms on metallic materials. NPJ Mater. Degrad. 6, 25 (2022).
Hall-Stoodley, L. et al. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 65, 127–145 (2012).
Trego, A. C., Mills, S. & Collins, G. Granular biofilms: function, application, and new trends as model microbial communities. Crit. Rev. Environ. Sci. Technol. 51, 1702–1725 (2021). Focused and informative review on types, functions and applications of microbial granules.
Zetsche, E.-M., Larsson, A. I., Iversen, M. H. & Ploug, H. Flow and diffusion around and within diatom aggregates: effects of aggregate composition and shape. Limnol. Oceanogr. 65, 1818–1833 (2020).
Wilén, B.-M., Liébana, R., Persson, F., Modin, O. & Hermansson, M. The mechanisms of granulation of activated sludge in wastewater treatment, its optimization, and impact on effluent quality. Appl. Microbiol. Biotechnol. 102, 5005–5020 (2018).
Bjarnsholt, T. et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558 (2009). First paper to show, using specific 16S molecular FISH probes, P. aeruginosa in biofilm aggregates in situ in the respiratory tract in individuals with cystic fibrosis.
Bjarnsholt, T. et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16, 2–10 (2008).
Gloag, E. S., Wozniak, D. J., Stoodley, P. & Hall-Stoodley, L. Mycobacterium abscessus biofilms have viscoelastic properties which may contribute to their recalcitrance in chronic pulmonary infections. Sci. Rep. 11, 5020 (2021).
Jensen, L. K. et al. Novel porcine model of implant-associated osteomyelitis: a comprehensive analysis of local, regional, and systemic response. J. Orthop. Res. 35, 2211–2221 (2017).
Li, C., Renz, N. & Trampuz, A. Management of periprosthetic joint infection. Hip Pelvis 30, 138–146 (2018).
Dudareva, M. et al. Sonication versus tissue sampling for diagnosis of prosthetic joint and other orthopedic device-related infections. J. Clin. Microbiol. https://doi.org/10.1128/JCM.00688-18 (2018).
Fux, C. A., Wilson, S. & Stoodley, P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186, 4486–4491 (2004). First paper demonstrating that aggregates are continually shed from biofilms and demonstrate biofilm tolerance to antibiotics.
Bay, L. et al. Universal dermal microbiome in human skin. mBio https://doi.org/10.1128/mBio.02945-19 (2020).
Burmolle, M. et al. Biofilms in chronic infections - a matter of opportunity - monospecies biofilms in multispecies infections. FEMS Immunol. Med. Microbiol. 59, 324–336 (2010).
Kim, D. et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc. Natl Acad. Sci. USA 117, 12375–12386 (2020).
Bowen, W. H., Burne, R. A., Wu, H. & Koo, H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242 (2018).
Ashrafi, M. et al. Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Sci. Rep. 8, 9431 (2018).
Salta, M., Wharton, J. A., Blache, Y., Stokes, K. R. & Briand, J.-F. Marine biofilms on artificial surfaces: structure and dynamics. Environ. Microbiol. 15, 2879–2893 (2013).
Dastgheyb, S. S. et al. Staphylococcal persistence due to biofilm formation in synovial fluid containing prophylactic cefazolin. Antimicrob. Agents Chemother. 59, 2122–2128 (2015).
Landry, R. M., An, D., Hupp, J. T., Singh, P. K. & Parsek, M. R. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol. Microbiol. 59, 142–151 (2006).
Singh, P. K. et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764 (2000).
Worlitzsch, D. et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325 (2002).
Pestrak, M. J. et al. Investigation of synovial fluid induced Staphylococcus aureus aggregate development and its impact on surface attachment and biofilm formation. PLoS ONE 15, e0231791 (2020).
Macias-Valcayo, A. et al. Synovial fluid mediated aggregation of clinical strains of four enterobacterial species. Adv. Exp. Med. Biol. https://doi.org/10.1007/5584_2020_573 (2020).
Bidossi, A., Bottagisio, M., Savadori, P. & De Vecchi, E. Identification and characterization of planktonic biofilm-like aggregates in infected synovial fluids from joint infections. Front. Microbiol. 11, 1368 (2020).
Kragh, K. N. et al. The inoculation method could impact the outcome of microbiological experiments. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.02264-17 (2018).
Schleheck, D. et al. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS ONE 4, e5513 (2009).
Haaber, J., Cohn, M. T., Frees, D., Andersen, T. J. & Ingmer, H. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PLoS ONE 7, e41075 (2012).
Kragh, K. N. et al. Inoculation method could impact the outcome of microbiological experiments. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.02264-17 (2017).
Hall-Stoodley, L. & Stoodley, P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13, 7–10 (2005).
Secor, P. R., Michaels, L. A., Ratjen, A., Jennings, L. K. & Singh, P. K. Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 115, 10780 (2018).
Secor, P. R., Michaels, L. A., Ratjen, A., Jennings, L. K. & Singh, P. K. Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 115, 10780–10785 (2018).
Dastgheyb, S., Parvizi, J., Shapiro, I. M., Hickok, N. J. & Otto, M. Effect of biofilms on recalcitrance of staphylococcal joint infection to antibiotic treatment. J. Infect. Dis. 211, 641–650 (2015). First paper demonstrating the importance of host factors in rapid aggregation of S. aureus into biofilm-like aggregates.
Knott, S. et al. Staphylococcus aureus floating biofilm formation and phenotype in synovial fluid depends on albumin, fibrinogen, and hyaluronic acid. Front. Microbiol. 12, 655873 (2021).
Dar, D., Dar, N., Cai, L. & Newman, D. K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science https://doi.org/10.1126/science.abi4882 (2021). Using an innovative transcriptome-imaging approach, this article provides visual evidence of differential gene expression and heterogeneity within biofilms at the single-cell level.
Dal Co, A., van Vliet, S. & Ackermann, M. Emergent microscale gradients give rise to metabolic cross-feeding and antibiotic tolerance in clonal bacterial populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190080 (2019).
Kowalski, C. H., Morelli, K. A., Schultz, D., Nadell, C. D. & Cramer, R. A. Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc. Natl Acad. Sci. USA 117, 22473–22483 (2020).
Cornforth, D. M. et al. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl Acad. Sci. USA 115, E5125–E5134 (2018).
Rossi, E., Falcone, M., Molin, S. & Johansen, H. K. High-resolution in situ transcriptomics of Pseudomonas aeruginosa unveils genotype independent patho-phenotypes in cystic fibrosis lungs. Nat. Commun. 9, 3459 (2018).
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P. & Hall-Stoodley, L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755 (2017).
Kragh, K. N. et al. Role of multicellular aggregates in biofilm formation. mBio 7, e00237 (2016).
Hawes, I., Sumner, D. & Jungblut, A. D. in The Structure and Function of Aquatic Microbial Communities (ed. Hurst, C. J.) 91–120 (Springer International Publishing, 2019).
Franca, R. D. G., Pinheiro, H. M., van Loosdrecht, M. C. M. & Lourenço, N. D. Stability of aerobic granules during long-term bioreactor operation. Biotechnol. Adv. 36, 228–246 (2018).
Li, Y. et al. Anaerobic microbiologically influenced corrosion mechanisms interpreted using bioenergetics and bioelectrochemistry: a review. J. Mater. Sci. Technol. 34, 1713–1718 (2018).
Bahrami, A., Khouzani, M. K. & Harchegani, B. B. Establishing the root cause of a failure in a firewater pipeline. Eng. Fail. Anal. 127, 105474 (2021).
Risse-Buhl, U. et al. The role of hydrodynamics in shaping the composition and architecture of epilithic biofilms in fluvial ecosystems. Water Res. 127, 211–222 (2017).
Kirketerp-Moller, K., Stewart, P. S. & Bjarnsholt, T. The zone model: a conceptual model for understanding the microenvironment of chronic wound infection. Wound Repair Regen. 28, 593–599 (2020).
Chan, C. S. et al. The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front. Microbiol. https://doi.org/10.3389/fmicb.2016.00796 (2016).
Bay, L. et al. Bacterial aggregates establish at the edges of acute epidermal wounds. Adv. Wound Care 7, 105–113 (2018).
Ring, H. C. et al. Bacterial biofilm in chronic lesions of hidradenitis suppurativa. Br. J. Dermatol. 176, 993–1000 (2017).
Ring, H. C. et al. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 153, 897–905 (2017).
Qvist, T. et al. Chronic pulmonary disease with Mycobacterium abscessus complex is a biofilm infection. Eur. Respir. J. 46, 1823–1826 (2015).
Folsom, J. P. et al. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 10, 294 (2010).
Alhede, M. et al. Bacterial aggregate size determines phagocytosis efficiency of polymorphonuclear leukocytes. Med. Microbiol. Immunol. 209, 669–680 (2020).
Pamp, S. J., Gjermansen, M., Johansen, H. K. & Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68, 223–240 (2008).
Díaz-Pascual, F. et al. Spatial alanine metabolism determines local growth dynamics of Escherichia coli colonies. eLife 10, e70794 (2021).
Bjarnsholt, T. et al. The impact of mental models on the treatment and research of chronic infections due to biofilms. APMIS https://doi.org/10.1111/apm.13163 (2021).
Kirketerp-Moller, K. et al. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46, 2717–2722 (2008).
Vasconcelos, C. et al. Lithifying microbial mats in Lagoa Vermelha, Brazil: modern Precambrian relics? Sediment. Geol. 185, 175–183 (2006).
Gilbertie, J. M. et al. Equine or porcine synovial fluid as a novel ex vivo model for the study of bacterial free-floating biofilms that form in human joint infections. PLoS ONE 14, e0221012 (2019).
Dorken, G., Ferguson, G. P., French, C. E. & Poon, W. C. K. Aggregation by depletion attraction in cultures of bacteria producing exopolysaccharide. J. R. Soc. Interface 9, 3490–3502 (2012). First paper linking polymer depletion with bacterial aggregates.
O’Toole, G. A. & Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304 (1998).
Schembri, M. A., Kjaergaard, K. & Klemm, P. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48, 253–267 (2003).
Whiteley, M. et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 (2001).
Wagner, V. E., Bushnell, D., Passador, L., Brooks, A. I. & Iglewski, B. H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095 (2003).
Bagge, N. et al. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob. Agents Chemother. 48, 1175–1187 (2004).
Ren, Y. et al. Emergent heterogeneous microenvironments in biofilms: substratum surface heterogeneity and bacterial adhesion force-sensing. FEMS Microbiol. Rev. 42, 259–272 (2018).
Serra, D. O. & Hengge, R. Stress responses go three dimensional – the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 16, 1455–1471 (2014).
Povolotsky, T. L., Keren-Paz, A. & Kolodkin-Gal, I. Metabolic microenvironments drive microbial differentiation and antibiotic resistance. Trends Genet. 37, 4–8 (2021).
Liao, J., Schurr, M. J. & Sauer, K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. J. Bacteriol. 195, 3352–3363 (2013).
Kowalski, C. H., Morelli, K. A., Stajich, J. E., Nadell, C. D. & Cramer, R. A. A heterogeneously expressed gene family modulates the biofilm architecture and hypoxic growth of Aspergillus fumigatus. mBio https://doi.org/10.1128/mBio.03579-20 (2021).
Nair, H. A., Periasamy, S., Yang, L., Kjelleberg, S. & Rice, S. A. Real time, spatial, and temporal mapping of the distribution of c-di-GMP during biofilm development. J. Biol. Chem. 292, 477–487 (2017).
Klauck, G., Serra, D. O., Possling, A. & Hengge, R. Spatial organization of different sigma factor activities and c-di-GMP signalling within the three-dimensional landscape of a bacterial biofilm. Open Biol. 8, 180066 (2018).
Lenz, A. P., Williamson, K. S., Pitts, B., Stewart, P. S. & Franklin, M. J. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 74, 4463–4471 (2008).
Shrout, J. D. et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62, 1264–1277 (2006).
Krasteva, P. V. et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327, 866–868 (2010).
Boles, B. R. & Horswill, A. R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052 (2008).
Rumbo-Feal, S. et al. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS ONE 8, e72968 (2013).
Bielecki, P. et al. In-vivo expression profiling of Pseudomonas aeruginosa infections reveals niche-specific and strain-independent transcriptional programs. PLoS ONE 6, e24235 (2011).
Potvin, E. et al. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5, 1294–1308 (2003).
Acknowledgements
The authors thank J. Story for help with preparing Fig. 3.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Ilana Kolodkin-Gal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Aggregates
-
Cohesive groups of microbial cells surrounded by extracellular polymeric substances and other entrapped abiotic or biotic materials. Microbial aggregates can be surface attached, matrix associated or free floating in the liquid phase and display a biofilm-like phenotype.
- Biofilms
-
Microbial aggregates attached or associated with a surface and embedded in a matrix. These can include single or multiple discrete aggregates or more continuous films.
- Aggregation
-
Any biological, chemical or physical process that enables microbial cells to form an aggregate.
- Growth
-
Expansion of aggregates by microbial growth and concomitant production of extracellular polymeric substance, whether in suspension or attached to a surface.
- Attachment
-
Suspended single cells or aggregates adhere to a host cellular surface or an abiotic surface, either directly to the substratum or to previously attached microbial cells or clusters.
- Detachment
-
An overarching term encompassing all phase transfer processes in which microbial cells and extracellular polymeric substances move from the surface-attached phase to a fluid-borne phase. This term is specific to surface-attached biofilms.
- Dispersal
-
Specifically connotes an active and biologically regulated release of microbial cells from a suspended or attached biofilm aggregate.
- Disaggregation
-
Aggregated cells, whether in suspension or associated with a surface, that shed smaller microbial aggregates or individual cells into the fluid phase.
- Accumulation
-
The net result of attachment, aggregation, growth, disaggregation and detachment processes that leads to expansion or shrinkage of a biofilm or aggregate.
- Removal
-
Implies the response to a mechanical, chemical or enzymatic intervention that causes attached aggregates or cells to be released from the surface.
- Polymer bridging
-
The aggregation of microbial cells in suspension caused by polymers that adhere to cell wall components forming bridging bonds between multiple cells.
- Sloughing
-
The release of coherent layers of surface-attached biofilm by adhesive failure (that is, at the biofilm–substratum interface), generally by fluid shear. This mechanism is specific to surface-attached biofilms.
- Co-aggregation
-
The formation of aggregates (also known as clumps) in suspension by bacteria of different species.
- Depletion aggregation
-
The formation of aggregates in suspension through a colloidal physics phenomenon that occurs when polymers in solution are of high enough concentration and molecular weight to initiate phase separation, ‘forcing’ microbial cells together.
- Auto-aggregation
-
The formation of aggregates (also known as clumps) in suspension by bacteria of the same species.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sauer, K., Stoodley, P., Goeres, D.M. et al. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat Rev Microbiol 20, 608–620 (2022). https://doi.org/10.1038/s41579-022-00767-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00767-0
This article is cited by
-
Biofilm colonization and succession in a full-scale partial nitritation-anammox moving bed biofilm reactor
Microbiome (2024)
-
Promastigote EPS secretion and haptomonad biofilm formation as evolutionary adaptations of trypanosomatid parasites for colonizing honeybee hosts
npj Biofilms and Microbiomes (2024)
-
Beneficial applications of biofilms
Nature Reviews Microbiology (2024)
-
NIR emissive Ru(II) metallacycle as a sonosensitizer/sonocatalyst assisting in vivo bacterial diagnosis and eradicating multidrug-resistant biofilms
Science China Chemistry (2024)
-
Antimicrobial activities of Agave fructans against multi-resistant and biofilm-producing Staphylococcus aureus isolated from bovine mastitis
Veterinary Research Communications (2024)