Abstract
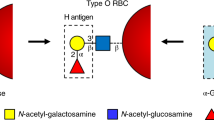
The 1013–1014 microorganisms present in the human gut (collectively known as the human gut microbiota) dedicate substantial percentages of their genomes to the degradation and uptake of carbohydrates, indicating the importance of this class of molecules. Carbohydrates function not only as a carbon source for these bacteria but also as a means of attachment to the host, and a barrier to infection of the host. In this Review, we focus on the diversity of carbohydrate-active enzymes (CAZymes), how gut microorganisms use them for carbohydrate degradation, the different chemical mechanisms of these CAZymes and the roles that these microorganisms and their CAZymes have in human health and disease. We also highlight examples of how enzymes from this treasure trove have been used in manipulation of the microbiota for improved health and treatment of disease, in remodelling the glycans on biopharmaceuticals and in the potential production of universal O-type donor blood.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L. & Gordon, J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Martens, E. C., Kelly, A. G., Tauzin, A. S. & Brumer, H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J. Mol. Biol. 426, 3851–3865 (2014).
Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R. & Goodman, A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019).
Johnson, K. V. A. & Foster, K. R. Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 16, 647–655 (2018).
Proctor, L. What’s next for the human microbiome? Nature 569, 623–625 (2019).
El Kaoutari, A., Armougom, F., Gordon, J. I., Raoult, D. & Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504 (2013).
Riley, L. W., Raphael, E. & Faerstein, E. Obesity in the United States — dysbiosis from exposure to low-dose antibiotics? Front. Public. Heal. 1, 69 (2013).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Bäckhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Price, K., Lewis, J., Wyatt, G. & Fenwick, G. Flatulence — causes, relation to diet and remedies. Nahrung 32, 609–626 (1988).
Lombard, V., Ramulu, H. G., Drula, E., Coutinho, P. M. & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014).
Zechel, D. L. & Withers, S. G. Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc. Chem. Res. 33, 11–18 (2000).
Jongkees, S. A. K. & Withers, S. G. Unusual enzymatic glycoside cleavage mechanisms. Acc. Chem. Res. 47, 226–235 (2014).
Armendáriz-Ruiz, M., Rodríguez-González, J. A., Camacho-Ruíz, R. M. & Mateos-Díaz, J. C. in Lipases and Phospholipases: Methods and Protocols (ed. Sandoval, G.) 39–68 (Humana Press, 2018).
Agostoni, M., Hangasky, J. A. & Marletta, M. A. Physiological and molecular understanding of bacterial polysaccharide monooxygenases. Microbiol. Mol. Biol. Rev. 81, e00015–e00017 (2017).
Vaaje-Kolstad, G. et al. Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J. Mol. Biol. 416, 239–254 (2012).
Lairson, L. L., Henrissat, B., Davies, G. J. & Withers, S. G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008).
Boraston, A. B., Bolam, D. N., Gilbert, H. J. & Davies, G. J. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004).
Davies, G. J., Gloster, T. M. & Henrissat, B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr. Opin. Struct. Biol. 15, 637–645 (2005).
Koshland, D. E. Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. 28, 416–436 (1953).
Berlemont, R. & Martiny, A. C. Glycoside hydrolases across environmental microbial communities. PLoS Comput. Biol. 12, e1005300 (2016).
Danby, P. M. & Withers, S. G. Advances in enzymatic glycoside synthesis. ACS Chem. Biol. 11, 1784–1794 (2016).
Nishimoto, M. & Kitaoka, M. Practical preparation of lacto-N-biose I, a candidate for the Bifidus factor in human milk. Biosci. Biotechnol. Biochem. 71, 2101–2104 (2007).
Liu, Q. P. Y. P. et al. Bacterial glycosidases for the production of universal red blood cells. Nat. Biotechnol. 25, 454–464 (2007).
Rahfeld, P. et al. An enzymatic pathway in the human gut microbiome that converts A to universal O type blood. Nat. Microbiol. 4, 1475–1485 (2019). This paper highlights how functional metagenomics can be applied for discovery of enzymes with therapeutic potential from within the gut microbiome.
Muegge, B. D. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2017).
Sonnenburg, E. D. & Sonnenburg, J. L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390 (2019).
Groussin, M. et al. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 14319 (2017).
Freter, R., Brickner, H., Botney, M., Cleven, D. & Aranki, A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect. Immun. 39, 676–685 (1983).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–109 (2011).
Tap, J. et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ. Microbiol. 17, 4954–4964 (2015).
Makki, K., Deehan, E. C., Walter, J. & Bäckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23, 705–715 (2018).
Vangay, P. et al. US immigration westernizes the human gut microbiome. Cell 175, 962–972.e10 (2018).
Smits, S. A. et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–805 (2017).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
De Filippis, F., Pellegrini, N., Laghi, L., Gobbetti, M. & Ercolini, D. Unusual sub-genus associations of faecal Prevotella and Bacteroides with specific dietary patterns. Microbiome 4, 57 (2016).
Zaramela, L. S. et al. Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates. Nat. Microbiol. 4, 2082–2089 (2019).
Leeming, E. R., Johnson, A. J., Spector, T. D. & Le Roy, C. I. Effect of diet on the gut microbiota: rethinking intervention duration. Nutrients 11, 2862 (2019).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Hehemann, J.-H., Kelly, A. G., Pudlo, N. A., Martens, E. C. & Boraston, A. B. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc. Natl Acad. Sci. USA 109, 19786–19791 (2012). This paper provides key insights into how new capabilities for carbohydrate degradation can be obtained by gut bacteria via horizontal gene transfer.
Pluvinage, B. et al. Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont. Nat. Commun. 9, 1043 (2018).
Hehemann, J. H. et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 (2010).
Cerqueira, F. M., Photenhauer, A. L., Pollet, R. M., Brown, H. A. & Koropatkin, N. M. Starch digestion by gut bacteria: crowdsourcing for carbs. Trends Microbiol. 28, 95–108 (2020).
Anderson, K. L. & Salyers, A. A. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J. Bacteriol. 171, 3192–3198 (1989). This seminal work describes the first characterization of Sus from B. thetaiotaomicron.
Anderson, K. L. & Salyers, A. A. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 171, 3199–3204 (1989).
Brown, H. A. & Koropatkin, N. M. Host glycan utilization within the Bacteroidetes Sus-like paradigm. Glycobiology 31, 697–706 (2021).
Cuskin, F. et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517, 165–169 (2015).
Luis, A. S. et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat. Microbiol. 3, 210–219 (2018).
Rakoff-Nahoum, S., Foster, K. R. & Comstock, L. E. The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016).
Rakoff-Nahoum, S., Coyne, M. J. & Comstock, L. E. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24, 40–49 (2014).
Cartmell, A. et al. A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation. Nat. Microbiol. 3, 1314–1326 (2018).
Sobala, L. F. et al. An epoxide intermediate in glycosidase catalysis. ACS Cent. Sci. 6, 760–770 (2020). This study provides key experimental evidence for a substrate-assisted mechanism proceeding through a 1,2-epoxide intermediate by GH99 family members, one of the most recently elucidated glycoside hydrolase mechanisms.
Valguarnera, E., Scott, N. E., Azimzadeh, P. & Feldman, M. F. Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in Bacteroides. mSphere 3, e00559-18 (2018).
Briggs, J. A., Grondin, J. M. & Brumer, H. Communal living: glycan utilization by the human gut microbiota. Environ. Microbiol. 23, 15–35 (2021).
Ndeh, D. et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70 (2017). This tour de force offers insights into the coordination of carbohydrate degradation by gut bacteria, identifies several new CAZy families and revises the structure of RG-II (one of the most complex dietary glycans known).
Tamura, K. et al. Molecular mechanism by which prominent human gut bacteroidetes utilize mixed-linkage β-glucans, major health-promoting cereal polysaccharides. Cell Rep. 21, 417–430 (2017).
Rogowski, A. et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat. Commun. 6, 7481 (2015).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Lapébie, P., Lombard, V., Drula, E., Terrapon, N. & Henrissat, B. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 10, 2043 (2019).
Laine, R. A. Invited commentary: A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 10 structures for a reducing hexasaccharide: the isomer barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology 4, 759–767 (1994).
Terrapon, N. et al. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res. 46, D677–D683 (2018).
Despres, J. et al. Xylan degradation by the human gut Bacteroides xylanisolvens XB1AT involves two distinct gene clusters that are linked at the transcriptional level. BMC Genomics 17, 326 (2016).
Corfield, A. P. The interaction of the gut microbiota with the mucus barrier in health and disease in human. Microorganisms 6, 78 (2018).
Martens, E. C., Neumann, M. & Desai, M. S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 16, 457–470 (2018).
Ficko-Blean, E. & Boraston, A. B. Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr. Opin. Struct. Biol. 22, 570–577 (2012).
Sonnenburg, J. L., Angenent, L. T. & Gordon, J. I. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5, 569–573 (2004).
Tailford, L. E., Crost, E. H., Kavanaugh, D. & Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 6, 81 (2015).
Briliūtė, J. et al. Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci. Nat. Microbiol. 4, 1571–1581 (2019). This paper provides insights into how B. thetaiotaomicron utilizes extensive enzymatic machinery to carry out the degradation of structural variants of complex N-glycans.
Rahfeld, P. et al. Prospecting for microbial α-N-acetylgalactosaminidases yields a new class of GH31 O-glycanase. J. Biol. Chem. 294, 16400–16415 (2019).
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008).
Pudlo, N. A. et al. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. MBio 6, e01282–15 (2015).
Derrien, M., Belzer, C. & de Vos, W. M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 106, 171–181 (2017).
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T. & Gordon, J. I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl Acad. Sci. USA 96, 9833–9838 (1999).
Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. A model of host–microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383 (1996).
Benjdia, A. & Berteau, O. Sulfatases and radical SAM enzymes: emerging themes in glycosaminoglycan metabolism and the human microbiota. Biochem. Soc. Trans. 44, 109–115 (2016).
Varki, A. Nothing in glycobiology makes sense, except in the light of evolution. Cell 126, 841–845 (2006).
Luis, A. S. et al. A single sulfatase is required to access colonic mucin by a gut bacterium. Nature 598, 332–337 (2021).
Crouch, L. I. et al. Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown. Nat. Commun. 11, 4017 (2020). This work first identifies surface-localized endolytic glycoside hydrolases acting on O-glycans in Bacteroides (as had been observed in other PULs but not against O-glycans) and as such provided insights into degradation of mucosal glycans by gut bacteria.
Noach, I. et al. Recognition of protein-linked glycans as a determinant of peptidase activity. Proc. Natl Acad. Sci. USA 114, E679–E688 (2017). This insightful work first illustrated the molecular basis of glycoprotease activity.
Shon, D. J. et al. An enzymatic toolkit for selective proteolysis, detection, and visualization of mucin-domain glycoproteins. Proc. Natl Acad. Sci. USA 117, 21299–21307 (2020).
Haurat, M. F. et al. The glycoprotease CpaA secreted by medically relevant Acinetobacter species targets multiple O-linked host glycoproteins. mBio 11, e02033–20 (2020).
Malaker, S. A. et al. The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl Acad. Sci. USA 116, 7278–7287 (2019).
Pluvinage, B. et al. Architecturally complex O-glycopeptidases are customized for mucin recognition and hydrolysis. Proc. Natl Acad. Sci. USA 118, e2019220118 (2021).
Hughes, G. W. et al. The MUC5B mucin polymer is dominated by repeating structural motifs and its topology is regulated by calcium and pH. Sci. Rep. 9, 17350 (2019).
Renzi, F. et al. Glycan-foraging systems reveal the adaptation of Capnocytophaga canimorsus to the dog mouth. mBio 6, e02507–e02514 (2015).
Reily, C., Stewart, T. J., Renfrow, M. B. & Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 (2019).
Fairbanks, A. J. The ENGases: versatile biocatalysts for the production of homogeneous N-linked glycopeptides and glycoproteins. Chem. Soc. Rev. 46, 5128–5146 (2017).
Knapp, S. et al. NAG-Thiazoline, an N-acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc. 118, 6804–6805 (1996).
Vocadlo, D. J. & Withers, S. G. Detailed comparative analysis of the catalytic mechanisms of β-N-acetylglucosaminidases from families 3 and 20 of glycoside hydrolases. Biochemistry 44, 12809–12818 (2005).
Trastoy, B. et al. Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides. Nat. Commun. 11, 899 (2020).
Nihira, T. et al. Discovery of β-1,4-d-mannosyl-N-acetyl-d-glucosamine phosphorylase involved in the metabolism of N-glycans. J. Biol. Chem. 288, 27366–27374 (2013).
Higgins, M. A. et al. N-Glycan degradation pathways in gut- and soil-dwelling Actinobacteria share common core genes. ACS Chem. Biol. 16, 701–711 (2021).
Cordeiro, R. L. et al. N-Glycan utilization by Bifidobacterium gut symbionts involves a specialist β-mannosidase. J. Mol. Biol. 431, 732–747 (2019).
Cartmell, A. et al. How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc. Natl Acad. Sci. USA 114, 7037–7042 (2017). This work illustrates how B. thetaiotaomicron tackles the degradation of host glycosaminoglycans with highly variable sulfation patterns, specifically heparin and heparan sulfate.
Lombard, V. et al. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444 (2010).
Nakamichi, Y., Mikami, B., Murata, K. & Hashimoto, W. Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin. J. Biol. Chem. 289, 4787–4797 (2014).
Martens, E. C. et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9, e1001221 (2011).
Raghavan, V., Lowe, E. C., Townsend, G. E., Bolam, D. N. & Groisman, E. A. Tuning transcription of nutrient utilization genes to catabolic rate promotes growth in a gut bacterium. Mol. Microbiol. 93, 1010–1025 (2014).
Ndeh, D. et al. Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus. Nat. Commun. 11, 646 (2020).
Pellock, S. J. & Redinbo, M. R. Glucuronides in the gut: sugar-driven symbioses between microbe and host. J. Biol. Chem. 292, 8569–8576 (2017).
Bolleddula, J. & Chowdhury, S. K. Carbon–carbon bond cleavage and formation reactions in drug metabolism and the role of metabolic enzymes. Drug Metab. Rev. 47, 534–557 (2015).
Wallace, B. D. et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010).
Kong, R. et al. Old drug new use — amoxapine and its metabolites as potent bacterial β-glucuronidase inhibitors for alleviating cancer drug toxicity. Clin. Cancer Res. 20, 3521–3530 (2014).
Pollet, R. M. et al. An atlas of β-glucuronidases in the human intestinal microbiome. Structure 25, 967–977.e5 (2017).
Ervin, S. M. et al. Targeting regorafenib-induced toxicity through inhibition of gut microbial β-glucuronidases. ACS Chem. Biol. 14, 2737–2744 (2019).
Creekmore, B. C. et al. Mouse gut microbiome-encoded β-glucuronidases identified using metagenome analysis guided by protein structure. mSystems 4, e00452-19 (2019).
Pellock, S. J. et al. Gut microbial β-glucuronidase inhibition via catalytic cycle interception. ACS Cent. Sci. 4, 868–879 (2018).
Ng, K. M. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013). This paper provides an excellent illustration of how pathogens exploit antibiotic-induced dysbiosis and intercept sugars left available by disrupted cross-feeding relationships for infection.
Ganesh, B. P., Klopfleisch, R., Loh, G. & Blaut, M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella typhimurium-infected gnotobiotic mice. PLoS ONE 8, e74963 (2013).
Collins, J. et al. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553, 291–294 (2018).
Collins, J., Danhof, H. & Britton, R. A. The role of trehalose in the global spread of epidemic Clostridium difficile. Gut Microbes 10, 204–209 (2019).
Hobbs, J. K., Pluvinage, B. & Boraston, A. B. Glycan-metabolizing enzymes in microbe–host interactions: the Streptococcus pneumoniae paradigm. FEBS Lett. 592, 3865–3897 (2018).
Vahey, M. D. & Fletcher, D. A. Influenza A virus surface proteins are organized to help penetrate host mucus. eLife 8, e43764 (2019).
Etzold, S. & Juge, N. Structural insights into bacterial recognition of intestinal mucins. Curr. Opin. Struct. Biol. 28, 23–31 (2014).
Järvå, M. A. et al. Trefoil factors share a lectin activity that defines their role in mucus. Nat. Commun. 11, 2265 (2020).
Reeves, E. P. et al. Helicobacter pylori lipopolysaccharide interacts with TFF1 in a pH-dependent manner. Gastroenterology 135, 2043–2054.e2 (2008).
Ficko-Blean, E., Stubbs, K. A., Nemirovsky, O., Vocadlo, D. J. & Boraston, A. B. Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc. Natl Acad. Sci. USA 105, 6560–6565 (2008).
Varki, A. Biological roles of glycans. Glycobiology 27, 3–49 (2017).
Bardoel, B. W. et al. Identification of an immunomodulating metalloprotease of Pseudomonas aeruginosa (IMPa). Cell. Microbiol. 14, 902–913 (2012).
Szabady, R. L., Lokuta, M. A., Walters, K. B., Huttenlocher, A. & Welch, R. A. Modulation of neutrophil function by a secreted mucinase of Escherichia coli O157∶H7. PLoS Pathog. 5, e1000320 (2009).
Allhorn, M., Olin, A. I., Nimmerjahn, F. & Collin, M. Human IgG/FcγR interactions are modulated by streptococcal IgG glycan hydrolysis. PLoS ONE 3, e1413 (2008).
Collin, M. et al. EndoS and SpeB from Streptococcus pyogenes inhibit immunoglobulin-mediated opsonophagocytosis. Infect. Immun. 70, 6646–6651 (2002).
Cao, Y., Rocha, E. R. & Smith, C. J. Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proc. Natl Acad. Sci. USA 111, 12901–12906 (2014).
Lindhout, T. et al. Site-specific enzymatic polysialylation of therapeutic proteins using bacterial enzymes. Proc. Natl Acad. Sci. USA 108, 7397–7402 (2011).
Geissner, A. et al. 7-Fluorosialyl glycosides are hydrolysis resistant but readily assembled by sialyltransferases providing easy access to more metabolically stable glycoproteins. ACS Cent. Sci. 7, 345–354 (2021).
Kightlinger, W., Warfel, K. F., DeLisa, M. P. & Jewett, M. C. Synthetic glycobiology: parts, systems, and applications. ACS Synth. Biol. 9, 1534–1562 (2020).
Napiórkowska, M., Boilevin, J., Darbre, T., Reymond, J.-L. & Locher, K. P. Structure of bacterial oligosaccharyltransferase PglB bound to a reactive LLO and an inhibitory peptide. Sci. Rep. 8, 16297 (2018).
Wacker, M. et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 (2002).
Feldman, M. F. et al. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl Acad. Sci. USA 102, 3016–3021 (2005).
Stark, J. C. et al. On-demand biomanufacturing of protective conjugate vaccines. Sci. Adv. 7, eabe9444 (2021).
Valderrama-Rincon, J. D. et al. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat. Chem. Biol. 8, 434–436 (2012).
Natarajan, A. et al. Engineering orthogonal human O-linked glycoprotein biosynthesis in bacteria. Nat. Chem. Biol. 16, 1062–1070 (2020).
Du, T. et al. A bacterial expression platform for production of therapeutic proteins containing human-like O-linked glycans. Cell Chem. Biol. 26, 203–212.e5 (2019).
Bindels, L. B., Delzenne, N. M., Cani, P. D. & Walter, J. Opinion: Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 12, 303–310 (2015).
Delannoy-Bruno, O. et al. Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans. Nature 595, 91–95 (2021).
Fushinobu, S. Unique sugar metabolic pathways of Bifidobacteria. Biosci. Biotechnol. Biochem. 74, 2374–2384 (2010).
Charbonneau, M. R. et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164, 859–871 (2016).
Xiao, J. Z. et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl. Environ. Microbiol. 76, 54–59 (2010).
Bych, K. et al. Production of HMOs using microbial hosts — from cell engineering to large scale production. Curr. Opin. Biotechnol. 56, 130–137 (2019).
Mackenzie, L. F., Wang, Q., Warren, R. A. J. & Withers, S. G. Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J. Am. Chem. Soc. 120, 5583–5584 (1998).
Schmölzer, K., Weingarten, M., Baldenius, K. & Nidetzky, B. Glycosynthase principle transformed into biocatalytic process technology: lacto-N-triose II production with engineered exo-hexosaminidase. ACS Catal. 9, 5503–5514 (2019).
Schmölzer, K., Weingarten, M., Baldenius, K. & Nidetzky, B. Lacto-N-tetraose synthesis by wild-type and glycosynthase variants of the β-N-hexosaminidase from Bifidobacterium bifidum. Org. Biomol. Chem. 17, 5661–5665 (2019).
Ruzic, L., Bolivar, J. M. & Nidetzky, B. Glycosynthase reaction meets the flow: continuous synthesis of lacto-N-triose II by engineered β-hexosaminidase immobilized on solid support. Biotechnol. Bioeng. 117, 1597–1602 (2020).
Noguchi, M., Tanaka, T., Gyakushi, H., Kobayashi, A. & Shoda, S. I. Efficient synthesis of sugar oxazolines from unprotected N-acetyl-2-amino sugars by using chloroformamidinium reagent in water. J. Org. Chem. 74, 2210–2212 (2009).
Sjögren, J., Lood, R. & Nägeli, A. On enzymatic remodeling of IgG glycosylation; unique tools with broad applications. Glycobiology 30, 254–267 (2020).
Li, C. et al. Site-selective chemoenzymatic modification on the core fucose of an antibody enhances its Fcγ receptor affinity and ADCC activity. J. Am. Chem. Soc. 143, 7828–7838 (2021). This recent example illustrates how modifications to the core fucose within N-glycans in the Fc domain can lead to desirable therapeutic properties.
Fairbanks, A. J. Chemoenzymatic synthesis of glycoproteins. Curr. Opin. Chem. Biol. 53, 9–15 (2019).
Parsons, T. B. et al. Optimal synthetic glycosylation of a therapeutic antibody. Angew. Chem. Int. Ed. 55, 2361–2367 (2016).
Liu, C.-P. et al. Glycoengineering of antibody (Herceptin) through yeast expression and in vitro enzymatic glycosylation. Proc. Natl Acad. Sci. USA 115, 720–725 (2018).
Huang, W., Giddens, J., Fan, S. Q., Toonstra, C. & Wang, L. X. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J. Am. Chem. Soc. 134, 12308–12318 (2012).
Giddens, J. P., Lomino, J. V., DiLillo, D. J., Ravetch, J. V. & Wang, L. X. Site-selective chemoenzymatic glycoengineering of Fab and Fc glycans of a therapeutic antibody. Proc. Natl Acad. Sci. USA 115, 12023–12027 (2018).
Tang, F. et al. Selective N-glycan editing on living cell surfaces to probe glycoconjugate function. Nat. Chem. Biol. 16, 766–775 (2020).
Lin, L. et al. Sequential glycosylation of proteins with substrate-specific N-glycosyltransferases. ACS Cent. Sci. 6, 144–154 (2020). This work brings the field closest to selective introduction of specific N-glycan structures to different sites within a protein — potentially enabling more thorough interrogation of the roles of different glycoforms.
Schwarz, F. et al. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat. Chem. Biol. 6, 264–266 (2010).
Xu, Y. et al. A novel enzymatic method for synthesis of glycopeptides carrying natural eukaryotic N-glycans. Chem. Commun. 53, 9075–9077 (2017).
Wardman, J. F. et al. Discovery and development of promiscuous O-glycan hydrolases for removal of intact sialyl T-antigen. ACS Chem. Biol. 16, 2004–2015 (2021).
Landsteiner, K. & Wiener, A. S. An agglutinable factor in human blood recognized by immune sera for rhesus blood. Exp. Biol. Med. 43, 223–223 (1940).
Daniels, G. & Reid, M. E. Blood groups: the past 50 years. Transfusion 50, 281–289 (2010).
Selleng, K. et al. Emergency transfusion of patients with unknown blood type with blood group O rhesus D positive red blood cell concentrates: a prospective, single-centre, observational study. Lancet Haematol. 4, e218–e224 (2017).
Goldstein, J., Siviglia, G., Hurst, R., Lenny, L. & Reich, L. Group B erythrocytes enzymatically converted to group O survive normally in A, B, and O individuals. Science 215, 168–170 (1982).
Yip, V. L. Y. et al. An unusual mechanism of glycoside hydrolysis involving redox and elimination steps by a family 4 β-glycosidase from Thermotoga maritima. J. Am. Chem. Soc. 126, 8354–8355 (2004).
Rossez, Y. et al. Almost all human gastric mucin O-glycans harbor blood group A, B or H antigens and are potential binding sites for Helicobacter pylori. Glycobiology 22, 1193–1206 (2012).
Tauzin, A. S. et al. Investigating host–microbiome interactions by droplet based microfluidics. Microbiome 8, 141 (2020).
Colin, P.-Y. et al. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 6, 10008 (2015).
Armstrong, Z. et al. High-throughput recovery and characterization of metagenome-derived glycoside hydrolase-containing clones as a resource for biocatalyst development. mSystems 4, e00082-19 (2019).
Wolfenden, R., Lu, X. & Young, G. Spontaneous hydrolysis of glycosides. J. Am. Chem. Soc. 120, 6814–6815 (1998).
McCarter, J. D. & Stephen Withers, G. Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4, 885–892 (1994).
Davies, G. et al. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology 28, 3–8 (2018).
Grondin, J. M., Tamura, K., Déjean, G., Abbott, D. W. & Brumer, H. Polysaccharide utilization loci: fueling microbial communities. J. Bacteriol. 199, e00860–e00816 (2017).
Glenwright, A. J. et al. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541, 407–411 (2017). This paper reports the first crystal structure of SusCD complexes from B. thetaiotaomicron and provides evidence for a ‘pedal bin’ mechanism.
Acknowledgements
The authors were supported by funding from the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council of Canada (NSERC) during the writing of this Review.
Author information
Authors and Affiliations
Contributions
All authors contributed to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
P.R. and S.G.W. are co-founders of a company to develop enzymes for conversion of blood type. The research underlying that is mentioned in this Review. J.F.W. and R.K.B. declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks David Bolam, Bernard Henrissat and Nicole Koropatkin for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
CAZy database: http://www.cazy.org/
CAZypedia: https://www.cazypedia.org
Glossary
- Short-chain fatty acids
-
Fatty acids containing up to six carbon atoms (predominantly acetate, propionate and butyrate in the human gut) that can be produced by gut microorganisms during fermentation.
- Cross-feeding
-
A phenomenon in which microorganisms utilize metabolites and degradation products derived from other microorganisms.
- Outer membrane vesicles
-
Vesicles with distinct protein cargo that are derived from the outer membrane of Gram-negative bacteria and are released into the environment.
- E1cb mechanism
-
A two-step chemical reaction in which an acidic proton is abstracted by a base to form a stabilized carbanion, followed by elimination of a leaving group as the carbanion lone pair moves to form a π-bond with the adjacent atoms.
- Xenobiotics
-
Molecules that are not produced or expected to be found within an organism (for example, drugs).
- Endobiotics
-
Molecules that are produced or are expected to be found within an organism (for example, hormones).
- Antibody-dependent cellular toxicity
-
An immune response in which binding of antibodies to a cell surface results in the lysis of that cell by effector cells of the immune system.
Rights and permissions
About this article
Cite this article
Wardman, J.F., Bains, R.K., Rahfeld, P. et al. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat Rev Microbiol 20, 542–556 (2022). https://doi.org/10.1038/s41579-022-00712-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00712-1
This article is cited by
-
Role of polyphenols in remodeling the host gut microbiota in polycystic ovary syndrome
Journal of Ovarian Research (2024)
-
Gut microbial CAZymes markers for depression
Translational Psychiatry (2024)
-
Simulated digestions of free oligosaccharides and mucin-type O-glycans reveal a potential role for Clostridium perfringens
Scientific Reports (2024)
-
Unveiling microbial guilds and symbiotic relationships in Antarctic sponge microbiomes
Scientific Reports (2024)
-
Relationships between diet and gut microbiome in an Italian and Dutch cohort: does the dietary protein to fiber ratio play a role?
European Journal of Nutrition (2024)