Abstract
Actinobacteria constitute a highly diverse bacterial phylum with an unrivalled metabolic versatility. They produce most of the clinically used antibiotics and a plethora of other natural products with medical or agricultural applications. Modern ‘omics’-based technologies have revealed that the genomic potential of Actinobacteria greatly outmatches the known chemical space. In this Review, we argue that combining insights into actinobacterial ecology with state-of-the-art computational approaches holds great promise to unlock this unexplored reservoir of actinobacterial metabolism. This enables the identification of small molecules and other stimuli that elicit the induction of poorly expressed biosynthetic gene clusters, which should help reinvigorate screening efforts for their precious bioactive natural products.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Whitman, W. B. et al. Bergey’s Manual of Systematic Bacteriology: Volume 5: The Actinobacteria (Springer Science & Business Media, 2012).
van der Meij, A., Worsley, S. F., Hutchings, M. I. & van Wezel, G. P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416 (2017).
Barka, E. A. et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43 (2016).
Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 58, 1–26 (2005).
Hopwood, D. A. Streptomyces in Nature and Medicine: the Antibiotic Makers (Oxford Univ. Press, 2007).
Vrancken, K. & Anne, J. Secretory production of recombinant proteins by Streptomyces. Future Microbiol. 4, 181–188 (2009).
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70, 461–477 (2007).
Davies, J. & Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 (2010).
Cooper, M. A. & Shlaes, D. Fix the antibiotics pipeline. Nature 472, 32 (2011).
Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug. Discov. 6, 29–40 (2007).
Baltz, R. H. Antimicrobials from actinomycetes: back to the future. Microbe 2, 125–131 (2007).
Baltz, R. H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8, 557–563 (2008).
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531 (2003).
van den Berg, M. A. et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 26, 1161–1168 (2008).
Bode, H. B., Bethe, B., Hofs, R. & Zeeck, A. Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem 3, 619–627 (2002).
Romano, S., Jackson, S. A., Patry, S. & Dobson, A. D. W. Extending the “One strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 16, 244 (2018).
Wu, C. et al. Lugdunomycin, an angucycline-derived molecule with unprecedented chemical architecture. Angew. Chem. Int. Ed. Engl. 58, 2809–2814 (2019).
Wright, E. S. & Vetsigian, K. H. Inhibitory interactions promote frequent bistability among competing bacteria. Nat. Commun. 7, 11274 (2016).
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010).
Abrudan, M. I. et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl Acad. Sci. USA 112, 11054–11059 (2015).
Traxler, M. F. & Kolter, R. Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 32, 956–970 (2015).
Traxler, M. F., Watrous, J. D., Alexandrov, T., Dorrestein, P. C. & Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4, e00459-13 (2013). This study uses nanospray desorption electrospray ionization mass spectrometry and MALDI–TOF imaging mass spectrometry to identify the chemical interactions that take place between actinomycete bacteria, revealing substantial changes in specialized metabolite production.
Rutledge, P. J. & Challis, G. L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523 (2015).
Zhu, H., Sandiford, S. K. & van Wezel, G. P. Triggers and cues that activate antibiotic production by actinomycetes. J. Ind. Microbiol. Biotechnol. 41, 371–386 (2014).
Guerinot, M. L. Microbial iron transport. Annu. Rev. Microbiol. 48, 743–772 (1994).
Li, C., Ji, C. & Tang, B. Purification, characterisation and biological activity of melanin from Streptomyces sp. FEMS Microbiol. Lett. 365, fny077 (2018).
Sadeghi, A. et al. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 169, 699–708 (2014).
Kroiss, J. et al. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6, 261–263 (2010).
Raaijmakers, J. M. & Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424 (2012).
Doroghazi, J. R. & Metcalf, W. W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics 14, 611 (2013).
Cimermancic, P. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014).
Sriswasdi, S., Yang, C. C. & Iwasaki, W. Generalist species drive microbial dispersion and evolution. Nat. Commun. 8, 1162 (2017).
Shimkets, L. J. in Bacterial Genomes: Physical Structure and Analysis (eds de Bruijn, F. J., Lupski, J. R. & Weinstock, G. M.) 5–11 (Chapman & Hall, 1998).
Fraser, C. M. et al. The minimal gene complement of Mycoplasma genitalium. Science 270, 397–403 (1995).
Salem, H. et al. Drastic genome reduction in an herbivore’s pectinolytic symbiont. Cell 171, 1520–1531 (2017).
Davies, J. Millennium bugs. Trends Cell Biol. 9, M2–M5 (1999).
Claessen, D., Rozen, D. E., Kuipers, O. P., Sogaard-Andersen, L. & van Wezel, G. P. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124 (2014).
Flärdh, K. & Buttner, M. J. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49 (2009).
Chater, K. F. & Losick, R. in Bacteria as Multicellular Organisms (eds Shapiro, J. A. & Dworkin, M.) 149–182 (Oxford University Press, 1997).
Merrick, M. J. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96, 299–315 (1976).
Hopwood, D. A., Wildermuth, H. & Palmer, H. M. Mutants of Streptomyces coelicolor defective in sporulation. J. Gen. Microbiol. 61, 397–408 (1970).
Bibb, M. J. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215 (2005).
van der Heul, H. U., Bilyk, B. L., McDowall, K. J., Seipke, R. F. & van Wezel, G. P. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat. Prod. Rep. 35, 575–604 (2018).
Manteca, A. A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent surface cultures of Streptomyces antibioticus. Microbiology 151, 3689–3697 (2005).
Manteca, A., Mader, U., Connolly, B. A. & Sanchez, J. A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics 6, 6008–6022 (2006).
Tenconi, E., Traxler, M. F., Hoebreck, C., van Wezel, G. P. & Rigali, S. Production of prodiginines is part of a programmed cell death process in Streptomyces coelicolor. Front. Microbiol. 9, 1742 (2018).
Ohnishi, Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008).
Gomez-Escribano, J. P. et al. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 3, 2716–2720 (2012).
Wu, C. et al. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci. Rep. 5, 10868 (2015).
Challis, G. L. & Hopwood, D. A. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl Acad. Sci. USA 100, 14555–14561 (2003).
Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015).
Wilson, M. C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014).
Seipke, R. F., Kaltenpoth, M. & Hutchings, M. I. Streptomyces as symbionts: an emerging and widespread theme? FEMS. Microbiol. Rev. 36, 862–876 (2012).
Jensen, P. R., Williams, P. G., Oh, D.-C., Zeigler, L. & Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Env. Microbiol. 73, 1146–1152 (2007).
Yang, A. et al. Nitrosporeusines A and B, unprecedented thioester-bearing alkaloids from the Arctic Streptomyces nitrosporeus. Org. Lett. 15, 5366–5369 (2013).
Sayed, A. M. et al. Extreme environments: microbiology leading to specialized metabolites. J. Appl. Microbiol. 128, 630–657 (2019).
Chevrette, M. G. et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 10, 516 (2019). This article describes the biosynthetic potential of insect-associated streptomycetes and the use of metabolomics for strain prioritization, resulting in the discovery of a novel antifungal agent active against multidrug-resistant fungal strains.
Zipperer, A. et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016).
Ziemert, N. et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl Acad. Sci. USA 111, E1130–E1139 (2014). This work shows high pathway diversity in recently diverged Salinispora spp., providing evidence for evolutionary drivers that allow bacteria to maximize their capacity to produce diverse secondary metabolites.
Adamek, M. et al. Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics 19, 426 (2018).
Andam, C. P., Choudoir, M. J., Vinh Nguyen, A., Sol Park, H. & Buckley, D. H. Contributions of ancestral inter-species recombination to the genetic diversity of extant Streptomyces lineages. ISME J. 10, 1731–1741 (2016).
Tidjani, A. et al. Massive gene flux drives genome diversity between sympatric Streptomyces conspecifics. mBio 10, e01533-19 (2019).
McDonald, B. R. & Currie, C. R. Lateral gene transfer dynamics in the ancient bacterial genus Streptomyces. mBio 8, e00644-17 (2017).
Medema, M. H., Cimermancic, P., Sali, A., Takano, E. & Fischbach, M. A. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comp. Biol. 10, e1004016 (2014).
Joynt, R. & Seipke, R. F. A phylogenetic and evolutionary analysis of antimycin biosynthesis. Microbiology 164, 28–39 (2018).
Chevrette, M. G. et al. Taxonomic and metabolic incongruence in the ancient genus Streptomyces. Front. Microbiol. 10, 2170 (2019).
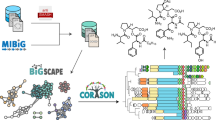
Navarro-Munoz, J. C. et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68 (2020). This work describes two computational tools, BiG-SCAPE and CORASON, designed to analyse BGC diversity in large genomic data sets.
Bruns, H. et al. Function-related replacement of bacterial siderophore pathways. ISME J. 12, 320–329 (2018). This study follows the evolutionary history of two siderophore biosynthesis pathways and reveals that acquisition of one pathway through HGT correlates to the loss of the other pathway.
Jensen, P. R. Natural products and the gene cluster revolution. Trends Microbiol. 24, 968–977 (2016).
Chater, K. F. & Chandra, G. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30, 651–672 (2006).
Ventura, M. et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548 (2007).
Choulet, F. et al. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol. Biol. Evol. 23, 2361–2369 (2006).
Bilyk, B., Horbal, L. & Luzhetskyy, A. Chromosomal position effect influences the heterologous expression of genes and biosynthetic gene clusters in Streptomyces albus J1074. Microb. Cell Fact. 16, 5 (2017).
Letzel, A.-C. et al. Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Env. Microbiol. 19, 3660–3673 (2017).
Ghinet, M. G. et al. Uncovering the prevalence and diversity of integrating conjugative elements in Actinobacteria. PLoS One 6, e27846 (2011).
Kinashi, H., Shimaji, M. & Sakai, A. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. Nature 328, 454–456 (1987).
Medema, M. H. et al. The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol. Evol. 2, 212–224 (2010).
Mochizuki, S. et al. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48, 1501–1510 (2003).
Hoskisson, P. A. & Fernández-Martínez, L. T. Regulation of specialised metabolites in Actinobacteria — expanding the paradigms. Env. Microbiol. Rep. 10, 231–238 (2018).
Huang, J. et al. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58, 1276–1287 (2005).
McLean, T. C., Hoskisson, P. A. & Seipke, R. F. Coordinate regulation of antimycin and candicidin biosynthesis. mSphere 1, e00305-16 (2016).
Martinet, L. et al. A single biosynthetic gene cluster is responsible for the production of bagremycin antibiotics and ferroverdin iron chelators. mBio 10, e01230-19 (2019).
Liu, G., Chater, K. F., Chandra, G., Niu, G. & Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143 (2013).
Wietzorrek, A. & Bibb, M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25, 1181–1184 (1997).
Autret, S., Nair, R. & Errington, J. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41, 743–755 (2001).
Gramajo, H. C., Takano, E. & Bibb, M. J. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7, 837–845 (1993).
Tomono, A., Tsai, Y., Yamazaki, H., Ohnishi, Y. & Horinouchi, S. Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 187, 5595–5604 (2005).
Lawlor, E. J., Baylis, H. A. & Chater, K. F. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1, 1305–1310 (1987).
Fernandez-Moreno, M. A., Caballero, J. L., Hopwood, D. A. & Malpartida, F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66, 769–780 (1991).
Takano, E. γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9, 287–294 (2006).
Willey, J. M. & Gaskell, A. A. Morphogenetic signaling molecules of the streptomycetes. Chem. Rev. 111, 174–187 (2011).
Tahlan, K. et al. Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 63, 951–961 (2007).
Wang, L. et al. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl Acad. Sci. USA 106, 8617–8622 (2009).
Willems, A. R. et al. Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. J. Mol. Biol. 376, 1377–1387 (2008).
Francis, I. M., Jourdan, S., Fanara, S., Loria, R. & Rigali, S. The cellobiose sensor CebR is the gatekeeper of Streptomyces scabies pathogenicity. mBio 6, e02018 (2015).
Rigali, S. et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by. Streptomyces. EMBO Rep. 9, 670–675 (2008). This work is the first to describe a complete signalling pathway from an extracellular nutrient (GlcNAc) to the control of antibiotic biosynthetic pathways.
Nazari, B. et al. Chitin-induced gene expression involved in secondary metabolic pathways in Streptomyces coelicolor A3(2) grown in soil. Appl. Env. Microbiol. 79, 707–713 (2012).
Craig, M. et al. Unsuspected control of siderophore production by N-acetylglucosamine in streptomycetes. Env. Microbiol. Rep. 4, 512–521 (2012).
Świątek-Połatyńska, M. A. et al. Genome-wide analysis of in vivo binding of the master regulator DasR in Streptomyces coelicolor identifies novel non-canonical targets. PLoS One 10, e0122479 (2015).
Urem, M., Swiatek-Polatynska, M. A., Rigali, S. & van Wezel, G. P. Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Mol. Microbiol. 102, 183–195 (2016).
Bode, H. Br., Bethe, B., Höfs, R. & Zeeck, A. Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem 3, 619–627 (2002).
Zhu, H. et al. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 160, 1714–1725 (2014).
Hosaka, T. et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 27, 462–464 (2009).
Tanaka, Y. et al. Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl. Env. Microbiol. 75, 4919–4922 (2009).
Bertrand, S. et al. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 32, 1180–1204 (2014).
Hoshino, S., Wakimoto, T., Onaka, H. & Abe, I. Chojalactones A–C, cytotoxic butanolides isolated from Streptomyces sp. cultivated with mycolic acid containing bacterium. Org. Lett. 17, 1501–1504 (2015).
Sugiyama, R. et al. 5-Alkyl-1,2,3,4-tetrahydroquinolines, new membrane-interacting lipophilic metabolites produced by combined culture of Streptomyces nigrescens and Tsukamurella pulmonis. Org. Lett. 17, 1918–1921 (2015).
Hsiao, N. H., Gottelt, M. & Takano, E. Chapter 6. Regulation of antibiotic production by bacterial hormones. Methods Enzymol. 458, 143–157 (2009).
Albright, J. C. et al. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol., 1535-1541 (2015).
Craney, A., Ozimok, C., Pimentel-Elardo, S. M., Capretta, A. & Nodwell, J. R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 19, 1020–1027 (2012). This work describes how screening a compound library for elicitors identified a novel family of molecules that activate antibiotic production.
Moon, K., Xu, F. & Seyedsayamdost, M. R. Cebulantin, a cryptic lanthipeptide antibiotic uncovered using bioactivity-coupled HiTES. Angew. Chem. Int. Ed. Engl. 58, 5973–5977 (2019).
Moon, K., Xu, F., Zhang, C. & Seyedsayamdost, M. R. Bioactivity-HiTES unveils cryptic antibiotics encoded in actinomycete bacteria. ACS Chem. Biol. 14, 767–774 (2019).
Kramer, J., Ozkaya, O. & Kummerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163 (2020).
Niu, G., Chater, K. F., Tian, Y., Zhang, J. & Tan, H. Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573 (2016).
Onaka, H., Mori, Y., Igarashi, Y. & Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Env. Microbiol. 77, 400–406 (2011).
Schroeckh, V. et al. Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl Acad. Sci. USA 106, 14558–14563 (2009).
Sung, A. A., Gromek, S. M. & Balunas, M. J. Upregulation and identification of antibiotic activity of a marine-derived Streptomyces sp. via co-cultures with human pathogens. Mar. Drugs 15 (2017).
Pérez, J. et al. Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microb. Biotechnol. 4, 175–183 (2011).
Patin, N. V., Floros, D. J., Hughes, C. C., Dorrestein, P. C. & Jensen, P. R. The role of inter-species interactions in Salinispora specialized metabolism. Microbiology 164, 946–955 (2018).
Ezaki, M. et al. Biphenomycin A production by a mixed culture. Appl. Environ. Microbiol. 58, 3879–3882 (1992).
Kurosawa, K. et al. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J. Am. Chem. Soc. 130, 1126–1127 (2008).
Traxler, M. F., Seyedsayamdost, M. R., Clardy, J. & Kolter, R. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol. Microbiol. 86, 628–644 (2012).
Onaka, H., Tabata, H., Igarashi, Y., Sato, Y. & Furumai, T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J. Antibiot. 54, 1036–1044 (2001).
Yang, Y. L., Xu, Y., Straight, P. & Dorrestein, P. C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 5, 885–887 (2009).
Mendes, R. et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 (2011).
Currie, C. R., Scott, J. A., Summerbell, R. C. & Malloch, D. D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398, 701–704 (1999).
Heine, D. et al. Chemical warfare between leafcutter ant symbionts and a co-evolved pathogen. Nat. Commun. 9, 2208 (2018). This work describes how Pseudonocardia, which protects a fungal garden against infection by a pathogenic fungus, elicits the production of counteracting antibiotics by the pathogen.
Spaepen, S. in Principles of Plant–Microbe Interactions (ed. Lugtenberg, B.) 247–256 (Springer, 2015).
van der Meij, A. et al. Inter- and intracellular colonization of Arabidopsis roots by endophytic Actinobacteria and the impact of plant hormones on their antimicrobial activity. Antonie Van. Leeuwenhoek 111, 679–690 (2018). This study describes the interaction between plants and streptomycetes, and the chemical interaction through which plants might influence specialized metabolite production by endophytic Actinobacteria.
Blin, K. et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). This work describes a computational tool used by researchers worldwide to predict biosynthetic gene clusters from genome sequences.
Skinnider, M. A., Merwin, N. J., Johnston, C. W. & Magarvey, N. A. PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 45, W49–W54 (2017). PRISM3 predicts various potential chemical products of biosynthetic gene clusters, demonstrating how the field is moving towards natural product structure prediction from a genomic sequence.
Kautsar, S. A. et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 48, D454–D458 (2019).
Doroghazi, J. R. et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014). This work shows how GCF classification can be used to mine large data sets for biosynthetic potential.
Alanjary, M. et al. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 45, W42–W48 (2017).
Schorn, M. A. et al. Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology 162, 2075–2086 (2016).
Charlop-Powers, Z. et al. Global biogeographic sampling of bacterial secondary metabolism. eLife 4, e05048 (2015).
Charlop-Powers, Z. et al. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proc. Natl Acad. Sci. USA 113, 14811–14816 (2016).
Tiwari, K. & Gupta, R. K. Rare actinomycetes: a potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 32, 108–132 (2012).
Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014).
Culp, E. J. et al. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat. Biotechnol. 37, 1149–1154 (2019). This work shows how genome engineering of Actinobacteria by removing genes for common antibiotics may result in the production of novel metabolites that previously remained unidentified.
Hiard, S. et al. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 357, 861–864 (2007).
Tan, K., Moreno-Hagelsieb, G., Collado-Vides, J. & Stormo, G. D. A comparative genomics approach to prediction of new members of regulons. Genome Res. 11, 566–584 (2001).
Rigali, S., Anderssen, S., Naome, A. & van Wezel, G. P. Cracking the regulatory code of biosynthetic gene clusters as a strategy for natural product discovery. Biochem. Pharmacol. 153, 24–34 (2018).
Carrion, V. J. et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612 (2019).
Duhrkop, K., Shen, H., Meusel, M., Rousu, J. & Bocker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl Acad. Sci. USA 112, 12580–12585 (2015).
van der Hooft, J. J., Wandy, J., Barrett, M. P., Burgess, K. E. & Rogers, S. Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl Acad. Sci. USA 113, 13738–13743 (2016).
Mohimani, H. et al. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 13, 30–37 (2017).
Mohimani, H. et al. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 9, 4035 (2018).
Ernst, M. et al. MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 9, 144 (2019).
Wilson, M. C. & Piel, J. Metagenomic approaches for exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. Chem. Biol. 20, 636–647 (2013).
Smanski, M. J. et al. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 14, 135–149 (2016).
Sugimoto, Y. et al. A metagenomic strategy for harnessing the chemical repertoire of the human microbiome. Science 366, eaax9176 (2019). This paper describes an innovative strategy to identify and characterize biosynthetic gene clusters from uncultivated microbes using metagenomics.
Smanski, M. J. et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 32, 1241–1249 (2014).
Shomar, H. et al. Metabolic engineering of a carbapenem antibiotic synthesis pathway in Escherichia coli. Nat. Chem. Biol. 14, 794–800 (2018).
Amos, G. C. A. et al. Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc. Natl Acad. Sci. USA 114, E11121–E11130 (2017).
Machado, H., Tuttle, R. N. & Jensen, P. R. Omics-based natural product discovery and the lexicon of genome mining. Curr. Opin. Microbiol. 39, 136–142 (2017).
Badri, D. V. & Vivanco, J. M. Regulation and function of root exudates. Plant. Cell Env. 32, 666–681 (2009).
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q. & Vivanco, J. M. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288, 4502–4512 (2013).
Bulgarelli, D. et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012).
Lebeis, S. L. et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864 (2015).
Culp, E. J. et al. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 578, 582–587 (2020).
van Heel, A. J. et al. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281 (2018).
Du, C. & van Wezel, G. P. Mining for microbial gems: integrating proteomics in the postgenomic natural product discovery pipeline. Proteomics 18, e1700332 (2018).
Goering, A. W. et al. Metabologenomics: correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2, 99–108 (2016).
Soldatou, S., Eldjarn, G. H., Huerta-Uribe, A., Rogers, S. & Duncan, K. R. Linking biosynthetic and chemical space to accelerate microbial secondary metabolite discovery. FEMS Microbiol. Lett. 366, fnz142 (2019).
Stokes, J. M. et al. A deep learning approach to antibiotic discovery. Cell 180, 688–702 (2020).
Acknowledgements
This work is supported by NACTAR grant 16440 from the Netherlands Organization for Scientific Research (NWO) to M.H.M. and G.P.v.W., and by the Profile area ‘Antibiotics’ of the Faculty of Sciences of Leiden University.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
M.H.M is a member of the Scientific Advisory Board of Hexagon Bio and co-founder of Design Pharmaceuticals. G.P.v.W. is in a committee for the Dutch Ministry of Health. D.A.v.B and B.R.T. do not declare any competing financial interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Siderophores
-
Chelating specialized metabolites produced by bacteria and fungi, which form a soluble complex with iron and thereby facilitate the uptake of this essential element.
- Endospore
-
A dormant, non-reproductive structure that can survive for long periods of time and is important for dispersal of the organism in the environment.
- Programmed cell death
-
(PCD). A cellular suicide mechanism found in eukaryotes and prokaryotes that is activated in response to different stimuli, including biotic and abiotic stresses.
- Vegetative hyphae
-
Long filamentous cells with multiple chromosomes that grow by tip extension and branching, forming a dense network into the surrounding substrate (often referred to as the vegetative or substrate mycelium).
- Sporogenic aerial hyphae
-
Reproductive hyphal structures that emerge from the vegetative mycelium into the air and develop into chains of unigenomic spores.
- Saprophytic bacteria
-
Bacteria, often found in the soil, that feed on dead and decaying organic material.
- Prodiginines
-
A family of red-pigmented, bacterial, specialized metabolites with a wide range of biological activities, including antibacterial, antitumour and immunosuppressive properties.
- Actinorhodin
-
A blue-pigmented polyketide antibiotic that is a model for polyketide biosynthesis.
- Ectoines
-
Bacterial specialized metabolites that act as osmolytes and protect against osmotic stress induced by, for example, changes in salt concentration of the environment.
- Terpenes
-
A class of specialized metabolites produced by plants, fungi and bacteria with different biological activities, often volatile and with a strong odour.
- Rhizosphere
-
The region of the soil and its microbial community immediately surrounding the plant root, which is directly influenced by root exudates.
Rights and permissions
About this article
Cite this article
van Bergeijk, D.A., Terlouw, B.R., Medema, M.H. et al. Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat Rev Microbiol 18, 546–558 (2020). https://doi.org/10.1038/s41579-020-0379-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-020-0379-y
This article is cited by
-
Effects of the alpine meadow in different phenological periods on rumen fermentation and gastrointestinal tract bacteria community in grazing yak on the Qinghai-Tibetan Plateau
BMC Microbiology (2024)
-
nanoRAPIDS as an analytical pipeline for the discovery of novel bioactive metabolites in complex culture extracts at the nanoscale
Communications Chemistry (2024)
-
Genomic and physiological characterization of Kitasatospora sp. nov., an actinobacterium with potential for biotechnological application isolated from Cerrado soil
Brazilian Journal of Microbiology (2024)
-
The sucrose non-fermenting-1 kinase Snf1 is involved in fludioxonil resistance via interacting with the high osmolarity glycerol MAPK kinase Hog1 in Fusarium
Phytopathology Research (2023)
-
A genomic catalogue of soil microbiomes boosts mining of biodiversity and genetic resources
Nature Communications (2023)