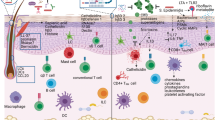
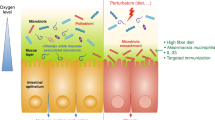
Abstract
Host–microbiota interactions are fundamental for the development of the immune system. Drastic changes in modern environments and lifestyles have led to an imbalance of this evolutionarily ancient process, coinciding with a steep rise in immune-mediated diseases such as autoimmune, allergic and chronic inflammatory disorders. There is an urgent need to better understand these diseases in the context of mucosal and skin microbiota. This Review discusses the mechanisms of how the microbiota contributes to the predisposition, initiation and perpetuation of immune-mediated diseases in the context of a genetically prone host. It is timely owing to the wealth of new studies that recently contributed to this field, ranging from metagenomic studies in humans and mechanistic studies of host–microorganism interactions in gnotobiotic models and in vitro systems, to molecular mechanisms with broader implications across immune-mediated diseases. We focus on the general principles, such as breaches in immune tolerance and barriers, leading to the promotion of immune-mediated diseases by gut, oral and skin microbiota. Lastly, the therapeutic avenues that either target the microbiota, the barrier surfaces or the host immune system to restore tolerance and homeostasis will be explored.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
01 June 2020
Fig. 2 file initially published online was corrupted and was replaced on 1/6/20.
References
Sansonetti, P. J. & Medzhitov, R. Learning tolerance while fighting ignorance. Cell 138, 416–420 (2009).
Lathrop, S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011).
Cebula, A. et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 497, 258–262 (2013).
Hand, T. W. et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556 (2012).
Gensollen, T., Iyer, S. S., Kasper, D. L. & Blumberg, R. S. How colonization by microbiota in early life shapes the immune system. Science 352, 539–544 (2016).
Ruff, W. E. & Kriegel, M. A. Autoimmune host-microbiota interactions at barrier sites and beyond. Trends Mol. Med. 21, 233–244 (2015).
Dehner, C., Fine, R. & Kriegel, M. A. The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr. Opin. Rheumatol. 31, 201–207 (2019).
Greiling, T. M. et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl Med. 10, eaan2306 (2018). This study demonstrated that orthologues of the primordial lupus autoantigen Ro60 expressed in human commensal bacteria can drive systemic autoimmunity by cross-reactivity with homologous regions in bacterial and host Ro60 protein.
Konig, M. F. et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl Med. 8, 369ra176 (2016). This study linked an oral pathobiont to post-translational modification of key autoantigens in rheumatoid arthritis. A toxin produced by this pathobiont induces both hypercitrullination of host proteins and activation of neutrophils, providing both innate and adaptive immune stimuli in the pathogenesis of rheumatoid arthritis.
Krebs, C. F. et al. Autoimmune renal disease is exacerbated by S1P-receptor-1-dependent intestinal Th17 cell migration to the kidney. Immunity 45, 1078–1092 (2016).
Manfredo Vieira, S. et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359, 1156–1161 (2018). This study found that E. gallinarum sequentially translocates to the mesenteric veins, lymph nodes, liver and spleen of autoimmune-prone animals, thereby driving organ-specific and systemic autoimmunity. The same species was recovered from livers of individuals with autoimmune diseases and promotes human hepatocyte activation as in mice.
Brubaker, S. W., Bonham, K. S., Zanoni, I. & Kagan, J. C. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 (2015).
Pancer, Z. & Cooper, M. D. The evolution of adaptive immunity. Annu. Rev. Immunol. 24, 497–518 (2006).
Iwasaki, A. & Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 (2015).
Chu, H. & Mazmanian, S. K. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675 (2013).
Bach, J. F. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat. Rev. Immunol. 18, 105–120 (2018).
von Mutius, E. & Vercelli, D. Farm living: effects on childhood asthma and allergy. Nat. Rev. Immunol. 10, 861–868 (2010).
Schroeder, B. O. & Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089 (2016).
Kundu, P., Blacher, E., Elinav, E. & Pettersson, S. Our gut microbiome: the evolving inner self. Cell 171, 1481–1493 (2017).
Sharon, G., Sampson, T. R., Geschwind, D. H. & Mazmanian, S. K. The central nervous system and the gut microbiome. Cell 167, 915–932 (2016).
Levy, M., Kolodziejczyk, A. A., Thaiss, C. A. & Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 17, 219–232 (2017).
Obata, T. et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl Acad. Sci. USA 107, 7419–7424 (2010).
Fung, T. C. et al. Lymphoid-tissue-resident commensal bacteria promote members of the IL-10 cytokine family to establish mutualism. Immunity 44, 634–646 (2016).
Klaasen, H. L. et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect. Immun. 61, 303–306 (1993).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Macpherson, A. J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004).
Gensollen, T. & Blumberg, R. S. Correlation between early-life regulation of the immune system by microbiota and allergy development. J. Allergy Clin. Immunol. 139, 1084–1091 (2017).
Renz, H. et al. The neonatal window of opportunity-early priming for life. J. Allergy Clin. Immunol. 141, 1212–1214 (2018).
Al Nabhani, Z. et al. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288 (2019).
Gomez de Aguero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012). This study provides experimental evidence for the hygiene hypothesis by demonstrating that neonatal natural killer T cells from germ-free mice promote IBD and asthma models in adulthood. Protection against gut and lung inflammation occurs by colonization with a microbiota during the neonatal period.
Livanos, A. E. et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 1, 16140 (2016).
Ruiz, V. E. et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat. Commun. 8, 518 (2017).
Scharschmidt, T. C. et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015).
Turner, J. R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 9, 799–809 (2009).
Luissint, A. C., Parkos, C. A. & Nusrat, A. Inflammation and the intestinal barrier: leukocyte-epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology 151, 616–632 (2016).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Bouskra, D. et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007).
Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). This study identified human IgA-coated pathobionts in IBD and functionally linked IgA-coated strains to colitis in a gnotobiotic model.
Castro-Dopico, T. et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity 50, 1099–1114 (2019). This study provides a previously underappreciated role for B cell responses against commensal bacteria in the pathogenesis of ulcerative colitis.
Stefka, A. T. et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl Acad. Sci. USA 111, 13145–13150 (2014).
Spadoni, I. et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science 350, 830–834 (2015).
Feehley, T. et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 25, 448–453 (2019). This study characterized protective human Clostridium spp. that mitigate cow’s milk food allergy in gnotobiotic animals.
Villasenor, A. & Stainier, D. Y. R. On the development of the hepatopancreatic ductal system. Semin. Cell Dev. Biol. 66, 69–80 (2017).
Tsuji, Y. et al. Sensing of commensal organisms by the intracellular sensor NOD1 mediates experimental pancreatitis. Immunity 37, 326–338 (2012). This study found that molecules from gut commensal bacteria promote autoimmune pancreatitis by triggering NOD1 receptors in vivo.
Costa, F. R. et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J. Exp. Med. 213, 1223–1239 (2016). This study found that gut commensal translocation and associated NOD2-activating molecules from commensal bacteria trigger murine type 1 diabetes. Together with Tsuji et al., these studies demonstrate that NOD1/2 ligands from the gut microbiota instigate autoimmunity in the pancreas (leading to either exocrine or endocrine inflammation).
Sorini, C. et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc. Natl Acad. Sci. USA 116, 15140–15149 (2019).
Balmer, M. L. et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci. Transl Med. 6, 237ra266 (2014).
Fine, R. L., Manfredo Vieira, S., Gilmore, M. S. & Kriegel, M. A. Mechanisms and consequences of gut commensal translocation in chronic diseases. Gut Microbes 11, 217–230 (2020).
Nakamoto, N. et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 4, 492–503 (2019). This study showed that K. pneumoniae, P. mirabilis and E. gallinarum are prevalent in microbiomes of individuals with ulcerative colitis and primary sclerosing cholangitis and translocate to mesenteric lymph nodes in gnotobiotic models, thereby promoting T H17-mediated autoimmunity.
Schwab, L. et al. Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance graft-versus-host disease via tissue damage. Nat. Med. 20, 648–654 (2014).
Fredricks, D. N. The gut microbiota and graft-versus-host disease. J. Clin. Invest. 129, 1808–1817 (2019).
Mathewson, N. D. et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 17, 505–513 (2016). This study demonstrated that the SCFA butyrate reduces intestinal epithelial cell injury in graft-versus-host disease, which is also mitigated by rationally selected butyrate-producing clostridial strains.
Johnson, K. V. & Foster, K. R. Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 16, 647–655 (2018).
Fung, T. C., Olson, C. A. & Hsiao, E. Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 20, 145–155 (2017).
Spadoni, I., Fornasa, G. & Rescigno, M. Organ-specific protection mediated by cooperation between vascular and epithelial barriers. Nat. Rev. Immunol. 17, 761–773 (2017).
Braniste, V. et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl Med. 6, 263ra158 (2014).
Erny, D. et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965–977 (2015).
Thion, M. S. et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell 172, 500–516 (2018).
Berer, K. et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479, 538–541 (2011). The murine gut microbiota, in particular SFB, were shown to trigger relapsing-remitting multiple sclerosis-like disease directed against the myelin oligodendrocyte glycoprotein autoantigen in gnotobiotic mice.
Rothhammer, V. et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016).
Berer, K. et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl Acad. Sci. USA 114, 10719–10724 (2017).
Cekanaviciute, E. et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl Acad. Sci. USA 114, 10713–10718 (2017).
Lamont, R. J., Koo, H. & Hajishengallis, G. The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759 (2018).
Dutzan, N. et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci. Transl Med. 10, eaat0797 (2018).
Hajishengallis, G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44 (2015).
Holers, V. M. et al. Rheumatoid arthritis and the mucosal origins hypothesis: protection turns to destruction. Nat. Rev. Rheumatol. 14, 542–557 (2018).
Mikuls, T. R. et al. Periodontitis and Porphyromonas gingivalis in patients with rheumatoid arthritis. Arthritis Rheumatol. 66, 1090–1100 (2014).
Segata, N. et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13, R42 (2012).
Atarashi, K. et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365 (2017).
Chen, B. et al. Proinflammatory and autoimmunogenic gut microbiome in systemic lupus erythematosus. bioRxiv https://doi.org/10.1101/621995 (2019).
Kemter, A. M. & Nagler, C. R. Influences on allergic mechanisms through gut, lung, and skin microbiome exposures. J. Clin. Invest. 130, 1483–1492 (2019).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012).
Gratz, I. K. et al. Cutting edge: memory regulatory t cells require IL-7 and not IL-2 for their maintenance in peripheral tissues. J. Immunol. 190, 4483–4487 (2013).
Scharschmidt, T. C. et al. Commensal microbes and hair follicle morphogenesis coordinately drive treg migration into neonatal skin. Cell Host Microbe 21, 467–477 (2017).
Chen, Y. E., Fischbach, M. A. & Belkaid, Y. Skin microbiota-host interactions. Nature 553, 427–436 (2018).
Bouslimani, A. et al. Molecular cartography of the human skin surface in 3D. Proc. Natl Acad. Sci. USA 112, E2120–E2129 (2015).
Naik, S. et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015).
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326 (2017).
Black, M., Bhattacharya, S., Philip, S., Norman, J. E. & McLernon, D. J. Planned cesarean delivery at term and adverse outcomes in childhood health. JAMA 314, 2271–2279 (2015).
Li, Y. et al. Cesarean delivery and risk of inflammatory bowel disease: a systematic review and meta-analysis. Scand. J. Gastroenterol. 49, 834–844 (2014).
Linehan, J. L. et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796 (2018).
Harrison, O. J. et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, eaat6280 (2019).
Byrd, A. L. et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl Med. 9, eaal4651 (2017). This study describes the strain diversity among Staphylococcus species in atopic dermatitis and linked clonal S. aureus strains with more severe disease. Cutaneous colonization of mice with S. aureus strains induced epidermal thickening and T H2 and T H17 infiltration in the skin.
Nakamura, Y. et al. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature 503, 397–401 (2013).
Kobayashi, T. et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 42, 756–766 (2015).
Nakatsuji, T. et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl Med. 9, eaah4680 (2017).
Williams, M. R. et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl Med. 11, eaat8329 (2019).
Nakatsuji, T. et al. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 4, 1431 (2013).
Nakatsuji, T. et al. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J. Invest. Dermatol. 136, 2192–2200 (2016).
Esaki, H. et al. Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J. Allergy Clin. Immunol. 138, 1639–1651 (2016).
Leonardi, S. et al. Serum interleukin 17, interleukin 23, and interleukin 10 values in children with atopic eczema/dermatitis syndrome (AEDS): association with clinical severity and phenotype. Allergy Asthma Proc. 36, 74–81 (2015).
Sparber, F. et al. The skin commensal yeast Malassezia triggers a type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation. Cell Host Microbe 25, 389–403 (2019).
Wolin, S. L. & Reinisch, K. M. The Ro 60 kDa autoantigen comes into focus: interpreting epitope mapping experiments on the basis of structure. Autoimmun. Rev. 5, 367–372 (2006).
Scholz, C. F. P. & Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 66, 4422–4432 (2016).
Dellacecca, E. R. et al. Antibiotics drive microbial imbalance and vitiligo development in mice. J. Invest. Dermatol. 140, 676–687 (2020).
Zanvit, P. et al. Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat. Commun. 6, 8424 (2015).
Kiyohara, H. et al. Toll-like receptor 7 agonist-induced dermatitis causes severe dextran sulfate sodium colitis by altering the gut microbiome and immune cells. Cell Mol. Gastroenterol. Hepatol. 7, 135–156 (2019).
Leyva-Castillo, J. M. et al. Mechanical skin injury promotes food anaphylaxis by driving intestinal mast cell expansion. Immunity 50, 1262–1275 (2019).
Man, W. H., de Steenhuijsen Piters, W. A. & Bogaert, D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270 (2017).
Marsland, B. J. & Gollwitzer, E. S. Host-microorganism interactions in lung diseases. Nat. Rev. Immunol. 14, 827–835 (2014).
Gollwitzer, E. S. et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 20, 642–647 (2014).
Palm, N. W., Rosenstein, R. K. & Medzhitov, R. Allergic host defences. Nature 484, 465–472 (2012).
Navarro, S. et al. Hookworm recombinant protein promotes regulatory T cell responses that suppress experimental asthma. Sci. Transl Med. 8, 362ra143 (2016).
Teo, S. M. et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe 24, 341–352 (2018).
Fujimura, K. E. et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191 (2016).
Arrieta, M. C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl Med. 7, 307ra152 (2015).
Trompette, A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166 (2014).
Thorburn, A. N. et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 6, 7320 (2015).
Roduit, C. et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 74, 799–809 (2019).
Dickson, R. P. et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 1, 16113 (2016).
Leonardi-Bee, J., Pritchard, D. & Britton, J. Asthma and current intestinal parasite infection: systematic review and meta-analysis. Am. J. Respir. Crit. Care Med. 174, 514–523 (2006).
Zaiss, M. M. et al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43, 998–1010 (2015).
Cantacessi, C. et al. Impact of experimental hookworm infection on the human gut microbiota. J. Infect. Dis. 210, 1431–1434 (2014).
Easton, A. V. et al. The impact of anthelmintic treatment on human gut microbiota based on cross-sectional and pre- and postdeworming comparisons in western Kenya. mBio 10, e00519-19 (2019).
Marson, A., Housley, W. J. & Hafler, D. A. Genetic basis of autoimmunity. J. Clin. Invest. 125, 2234–2241 (2015).
Gray, D. H., Gavanescu, I., Benoist, C. & Mathis, D. Danger-free autoimmune disease in Aire-deficient mice. Proc. Natl Acad. Sci. USA 104, 18193–18198 (2007).
Turer, E. E. et al. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J. Exp. Med. 205, 451–464 (2008).
Chu, H. et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016).
Taurog, J. D. et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 180, 2359–2364 (1994).
Silverman, M. et al. Protective major histocompatibility complex allele prevents type 1 diabetes by shaping the intestinal microbiota early in ontogeny. Proc. Natl Acad. Sci. USA 114, 9671–9676 (2017).
Zegarra-Ruiz, D. F. et al. A diet-sensitive commensal Lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe 25, 113–127 (2019). A resistant starch diet was shown to reduce an interferon-promoting L. reuteri strain in lupus models by fermentation to SCFAs that in turn suppress growth of the pathobiont. SCFAs also tightened the gut barrier, thereby preventing translocation of this strain to internal organs.
Proietti, M. et al. ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer’s patches to promote host-microbiota mutualism. Immunity 41, 789–801 (2014).
Faliti, C. E. et al. P2X7 receptor restrains pathogenic Tfh cell generation in systemic lupus erythematosus. J. Exp. Med. 216, 317–336 (2019).
Mukherjee, A. et al. Rheumatoid arthritis-associated autoimmunity due to Aggregatibacter actinomycetemcomitans and its resolution with antibiotic therapy. Front. Immunol. 9, 2352 (2018).
Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R. & Goodman, A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019).
Clayton, T. A., Baker, D., Lindon, J. C., Everett, J. R. & Nicholson, J. K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009).
Ben-Zvi, I., Kivity, S., Langevitz, P. & Shoenfeld, Y. Hydroxychloroquine: from malaria to autoimmunity. Clin. Rev. Allergy Immunol. 42, 145–153 (2012).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009).
Wang, C. et al. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell 163, 1413–1427 (2015).
Marino, E. et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 18, 552–562 (2017). The SCFAs butyrate and acetate increased T reg cells, decreased autoreactive T cells and strengthened the gut barrier in a model of T1D, supporting microbiota-derived diets as a therapeutic approach for autoimmune diseases.
Kawashima, T. et al. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-beta. Immunity 38, 1187–1197 (2013).
Kawashima, T. et al. Double-stranded RNA derived from lactic acid bacteria augments Th1 immunity via interferon-beta from human dendritic cells. Front. Immunol. 9, 27 (2018).
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L. & Gordon, J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Dahan, S., Segal, Y. & Shoenfeld, Y. Dietary factors in rheumatic autoimmune diseases: a recipe for therapy? Nat. Rev. Rheumatol. 13, 348–358 (2017).
Wu, C. et al. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517 (2013).
Kleinewietfeld, M. et al. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 496, 518–522 (2013). This study, together with Wu et al., links molecularly a high-salt diet with the induction of T H17 cells that promote multiple sclerosis and hypertension. The findings connect western dietary habits with non-gut metabolic and autoimmune diseases via the microbiota.
Wilck, N. et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551, 585–589 (2017).
Felton, S. et al. Ultraviolet radiation-induced upregulation of antimicrobial proteins in health and disease. Photochem. Photobiol. Sci. 12, 29–36 (2013).
Sim, S. & Wolin, S. L. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip. Rev. RNA 2, 686–699 (2011).
Catrina, A. I., Ytterberg, A. J., Reynisdottir, G., Malmstrom, V. & Klareskog, L. Lungs, joints and immunity against citrullinated proteins in rheumatoid arthritis. Nat. Rev. Rheumatol. 10, 645–653 (2014).
Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016). This study dissected microbiomes from geographically related infants predisposed to T1D and found that LPS from E. coli but not from B. dorei protects from autoimmunity in mice, supporting early immune education as a critical event.
Markle, J. G. et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088 (2013).
Yurkovetskiy, L. et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412 (2013).
Theofilopoulos, A. N., Kono, D. H. & Baccala, R. The multiple pathways to autoimmunity. Nat. Immunol. 18, 716–724 (2017).
Billi, A. C., Kahlenberg, J. M. & Gudjonsson, J. E. Sex bias in autoimmunity. Curr. Opin. Rheumatol. 31, 53–61 (2019).
Medzhitov, R., Schneider, D. S. & Soares, M. P. Disease tolerance as a defense strategy. Science 335, 936–941 (2012).
Torchinsky, M. B., Garaude, J., Martin, A. P. & Blander, J. M. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature 458, 78–82 (2009).
Campisi, L. et al. Apoptosis in response to microbial infection induces autoreactive TH17 cells. Nat. Immunol. 17, 1084–1092 (2016).
Omenetti, S. et al. The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity 51, 77–89 (2019).
Ansaldo, E. et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019).
Sanderson, N. S. et al. Cocapture of cognate and bystander antigens can activate autoreactive B cells. Proc. Natl Acad. Sci. USA 114, 734–739 (2017).
Munz, C., Lunemann, J. D., Getts, M. T. & Miller, S. D. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat. Rev. Immunol. 9, 246–258 (2009).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 (2008).
Huang, Z. et al. Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol. 4, 766–773 (2019). This study identified a monoclonal antibody against translocated peptidoglycan, a NOD2 ligand, that is elevated in human autoimmune disease. The antibody was shown to mitigate arthritis and multiple sclerosis-like disease in animal models.
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Horai, R. et al. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity 43, 343–353 (2015).
Tai, N. et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J. Exp. Med. 213, 2129–2146 (2016).
Hebbandi Nanjundappa, R. et al. A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell 171, 655–667 (2017). This study showed that a cross-reactive immune response against a pancreatic autoantigen can suppress inflammation in the gut.
Planas, R. et al. GDP-l-fucose synthase is a CD4+ T cell-specific autoantigen in DRB3*02:02 patients with multiple sclerosis. Sci. Transl Med. 10, eaat4301 (2018).
Ruff, W. E. et al. Pathogenic autoreactive T and B cells cross-react with mimotopes expressed by a common human gut commensal to trigger autoimmunity. Cell Host Microbe 26, 100–113 (2019). This study demonstrated cross-reactivity between Roseburia intestinalis at the molecular level and the key epitopes in a major autoantigen targeted by individuals with the autoimmune clotting disorder antiphospholipid syndrome. Human cross-reactivity was also linked to pathogenic effects in mice.
Gil-Cruz, C. et al. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science 366, 881–886 (2019).
Culina, S. et al. Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci. Immunol. 3, eaao4013 (2018).
Azzouz, D. et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 78, 947–956 (2019).
Bradley, C. P. et al. Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 22, 697–704 (2017).
Viladomiu, M. et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci. Transl Med. 9, eaaf9655 (2017). An adherent invasive E. coli strain is preferentially IgA coated in individuals with IBD and spondyloarthritis. This strain was further shown to drive antigen-specific T H17 cells and mediate colitis and arthritis in gnotobiotic animal models.
Zmora, N. et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405 (2018).
Smillie, C. S. et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23, 229–240 (2018).
Guo, C. J. et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell 168, 517–526 (2017).
Chen, H. et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell 177, 1217–1231 (2019).
Suez, J. & Elinav, E. The path towards microbiome-based metabolite treatment. Nat. Microbiol. 2, 17075 (2017).
Skelly, A. N., Sato, Y., Kearney, S. & Honda, K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 19, 305–323 (2019).
Garrett, W. S. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300 (2010).
Sheth, R. U., Cabral, V., Chen, S. P. & Wang, H. H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200 (2016).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Griffin, N. W. et al. Prior dietary practices and connections to a human gut microbial metacommunity alter responses to diet interventions. Cell Host Microbe 21, 84–96 (2017).
Bashiardes, S., Godneva, A., Elinav, E. & Segal, E. Towards utilization of the human genome and microbiome for personalized nutrition. Curr. Opin. Biotechnol. 51, 57–63 (2018).
Hashimoto, T. et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487, 477–481 (2012).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Kortright, K. E., Chan, B. K., Koff, J. L. & Turner, P. E. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232 (2019).
Duan, Y. et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511 (2019).
Nash, A. K. et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome 5, 153 (2017).
Lai, G. C., Tan, T. G. & Pavelka, N. The mammalian mycobiome: a complex system in a dynamic relationship with the host. Wiley Interdiscip. Rev. Syst. Biol. Med. 11, e1438 (2019).
Chiaro, T. R. et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl Med. 9, eaaf9044 (2017).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
McGovern, D. P. et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 42, 332–337 (2010).
Iliev, I. D. et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317 (2012). This study linked the mycobiome in IBD with inflammation mediated through Dectin-1, and identified polymorphisms in the gene encoding Dectin-1 of patients with ulcerative colitis.
Limon, J. J. et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377–388 (2019).
Yoshitomi, H. et al. A role for fungal {beta}-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J. Exp. Med. 201, 949–960 (2005).
Ruutu, M. et al. β-glucan triggers spondylarthritis and Crohn’s disease-like ileitis in SKG mice. Arthritis Rheum. 64, 2211–2222 (2012).
Wheeler, M. L. et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19, 865–873 (2016).
Li, X. et al. Response to fungal dysbiosis by gut-resident CX3CR1+ mononuclear phagocytes aggravates allergic airway disease. Cell Host Microbe 24, 847–856 (2018).
Bacher, P. et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against candida albicans. Cell 176, 1340–1355 (2019).
Virgin, H. W. The virome in mammalian physiology and disease. Cell 157, 142–150 (2014).
Neil, J. A. & Cadwell, K. The intestinal virome and immunity. J. Immunol. 201, 1615–1624 (2018).
Pfeiffer, J. K. & Virgin, H. W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351, aad5872 (2016).
Strickley, J. D. et al. Immunity to commensal papillomaviruses protects against skin cancer. Nature 575, 519–522 (2019).
Edwards, M. R. et al. Viral infections in allergy and immunology: how allergic inflammation influences viral infections and illness. J. Allergy Clin. Immunol. 140, 909–920 (2017).
Cadwell, K. et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010). This study provides an example of transkingdom interactions between a virus, the gut microbiota and host cell inflammation related to Crohn’s disease on the basis of a genetic predisposition.
Basic, M. et al. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm. Bowel Dis. 20, 431–443 (2014).
Oh, J. H. et al. Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont lactobacillus reuteri. Cell Host Microbe 25, 273–284 (2019).
Treger, R. S. et al. The lupus susceptibility locus Sgp3 encodes the suppressor of endogenous retrovirus expression SNERV. Immunity 50, 334–347 (2019).
Tabata, N. et al. Establishment of monoclonal anti-retroviral gp70 autoantibodies from MRL/lpr lupus mice and induction of glomerular gp70 deposition and pathology by transfer into non-autoimmune mice. J. Virol. 74, 4116–4126 (2000).
Young, G. R. et al. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491, 774–778 (2012).
Zeng, M. Y. et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658 (2016).
Maeda, Y. et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 68, 2646–2661 (2016).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013).
Pianta, A. et al. Evidence of the immune relevance of prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 69, 964–975 (2017).
Wu, H. J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Marietta, E. V. et al. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol. 68, 2878–2888 (2016).
Szymula, A. et al. T cell epitope mimicry between Sjogren’s syndrome antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin. Immunol. 152, 1–9 (2014).
Asahi, A. et al. Helicobacter pylori eradication shifts monocyte Fcγ receptor balance toward inhibitory FcγRIIB in immune thrombocytopenic purpura patients. J. Clin. Invest. 118, 2939–2949 (2008).
Jangi, S. et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015 (2016).
Lee, Y. K., Menezes, J. S., Umesaki, Y. & Mazmanian, S. K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4615–4622 (2011).
Cignarella, F. et al. Intermittent fasting confers protection in CNS autoimmunity by altering the gut microbiota. Cell Metab. 27, 1222–1235 (2018).
Ochoa-Reparaz, J. et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 185, 4101–4108 (2010).
Vatanen, T. et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018).
Mattner, J. et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe 3, 304–315 (2008).
McCarthy, D. D. et al. Mice overexpressing BAFF develop a commensal flora-dependent, IgA-associated nephropathy. J. Clin. Invest. 121, 3991–4002 (2011).
Laute-Caly, D. L. et al. The flagellin of candidate live biotherapeutic Enterococcus gallinarum MRx0518 is a potent immunostimulant. Sci. Rep. 9, 801 (2019).
Turley, S. J., Lee, J. W., Dutton-Swain, N., Mathis, D. & Benoist, C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc. Natl Acad. Sci. USA 102, 17729–17733 (2005).
Acknowledgements
The M.A.K. laboratory received funding from the National Institutes of Health (NIH) (K08AI095318, R01AI118855, T32AI07019, T32DK007017-39), the O’Brien Center at Yale (NIH P30DK079310), the Yale Rheumatic Diseases Research Core (NIH P30 AR053495), the Yale Liver Center (NIH P30 DK34989), the Women’s Health Research at Yale, the Arthritis National Research Foundation, the Arthritis Foundation, and the Lupus Research Institute.
Author information
Authors and Affiliations
Contributions
W.E.R, T.M.G. and M.A.K. equally conceived, wrote and edited the article. M.A.K. drafted the figures with input from W.E.R and T.M.G., and all authors contributed substantially to all aspects of the article and revised versions.
Corresponding author
Ethics declarations
Competing interests
M.A.K. received salary, consulting fees, honoraria or research funds from Roche, Bristol–Meyers Squibb, AbbVie and Cell Applications, and is an employee of Roche. M.A.K. holds a patent on the use of antibiotics and commensal vaccination to treat autoimmunity and received royalties. T.M.G. received research funds from Eli Lilly and Company and Janssen Scientific Affairs. W.E.R. declares no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks C. Nagler and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Pattern recognition receptors
-
Critical detection system of innate immunity, with different classes capable of recognizing a variety of evolutionarily conserved microorganism-associated molecular patterns.
- Toll-like receptors
-
(TLRs). Evolutionarily conserved pattern recognition receptors recognizing surface and endosomal ligands.
- Innate immunity
-
First line of defence that involves physical, chemical and various epithelial and innate immune responses that are rapid (within hours) and focused on detecting evolutionarily conserved patterns.
- Adaptive immunity
-
Second line of defence by lymphocytes using somatically rearranged receptors that can recognize any antigen and are initially slow (within days) before immunological memory is formed.
- Peyer’s patches
-
Anatomical structures in the small intestine composed of aggregated lymphoid follicles that act as immune sensors within the gut-associated lymphoid tissue.
- Secretory IgA
-
Main subclass of immunoglobulin found in mucus secretions, typically in dimeric form.
- Mesenteric lymph nodes
-
(MLNs). Organized secondary lymphoid organs within the mesentery that contain all immune components of peripheral lymph nodes but are receiving the draining lymph from the gut.
- Antimicrobial peptides
-
Secreted small molecules at barrier surfaces that have antimicrobial activity to contain the microbiota and pathogens.
- Nucleotide-binding oligomerization domain (NOD)-like receptors
-
Evolutionarily conserved pattern recognition receptors sensing intracellular ligands such as bacterial peptidoglycan.
- Inflammatory bowel disease
-
(IBD). A disease spectrum of ulcerative colitis and Crohn’s disease affecting the large or small bowels due to excessive immune responses towards the gut microbiota, gut epithelium and surrounding tissue.
- Type 1 diabetes
-
(T1D). Organ-specific autoimmune disease characterized by destruction of the endocrine pancreas, specifically the insulin-producing β-islet cells required for glucose homeostasis.
- Autoimmune pancreatitis
-
Chronic, organ-specific autoimmune disease due to autoimmune attack of the exocrine tissue of the pancreas and associated with elevated serum IgG4.
- Systemic lupus erythematosus
-
(SLE). Chronic, systemic autoimmune syndrome affecting the kidneys, skin, joints, brain and other organs but rarely involving the gut.
- Autoimmune hepatitis
-
Chronic organ-specific autoimmune disease of the liver that leads to immune cell-mediated hepatocyte damage and is associated with circulating autoantibodies.
- Gnotobiotic
-
An environment for rearing or culturing organisms in which all microorganisms are either known or excluded (germ free).
- Primary sclerosing cholangitis
-
(PSC). Chronic autoimmune disease destroying the bile ducts outside and within the liver, frequently associated with inflammatory bowel disease.
- Pathobionts
-
All potentially pathogenic microorganisms in a microbiota that are harmless under homeostasis but can provoke non-infectious diseases in a predisposed host; to be differentiated from opportunistic pathogens that cause infectious diseases in a predisposed host.
- Graft-versus-host disease
-
Acute or chronic alloimmune attack of host tissues by immune cells from the donor (graft). Graft-versus-host disease occurs after allogeneic bone marrow or stem cell transplantation.
- Rheumatoid arthritis
-
Systemic, rheumatic autoimmune disease causing primarily widespread inflammation of the joint linings (synovium) that leads to cartilage and bone destruction of small and large joints. It can also affect internal organs, such as the lung, leading to interstitial lung disease.
- Atopic dermatitis
-
Cutaneous inflammatory disease characterized by chronic relapsing pruritic skin rashes mediated by cutaneous barrier dysfunction and T helper 2 cell-skewed immune dysregulation.
- Sjögren’s syndrome
-
Chronic autoimmune disease targeting the salivary and lacrimal glands that leads to dryness of the eyes and mouth as well as systemic organ involvement and increased risk of lymphoma.
- Glomerular immune complex deposition
-
Deposition of autoantibody–autoantigen complexes in the glomeruli of the kidney that can lead to kidney damage and functional impairment.
- Lupus nephritis
-
Inflammation of the kidney in systemic lupus erythematosus that results from glomerular immune complex deposition and inflammation in the interstitium.
- Subacute cutaneous lupus erythematosus
-
A subtype of lupus that affects the skin, which can be associated with systemic lupus and is mediated by lymphocyte infiltration and immune complexes deposited in the skin.
- Vitiligo
-
A T cell-mediated skin autoimmune disease that manifests after loss of cutaneous melanocytes owing to T cell-mediated destruction with subsequent depigmentation of the skin.
- Multiple sclerosis
-
Chronic progressive or relapsing autoimmune neurologic disease due to immune-mediated destruction of myelin sheaths causing central nervous system dysfunction of varying severity.
- Mimotopes
-
Peptide sequences that mimic the structure of an epitope of a protein, carbohydrate or lipid antigen; a mimotope of a protein autoantigen can mimic a linear or conformational stretch of amino acids.
- Spondyloarthritis
-
A spectrum of rheumatic diseases that include ankylosing spondylitis and primarily affect the spine, leading to fusion or ankylosis. Spondyloarthritis can also involve peripheral joints, skin, gut and eyes.
Rights and permissions
About this article
Cite this article
Ruff, W.E., Greiling, T.M. & Kriegel, M.A. Host–microbiota interactions in immune-mediated diseases. Nat Rev Microbiol 18, 521–538 (2020). https://doi.org/10.1038/s41579-020-0367-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-020-0367-2
This article is cited by
-
Characteristics of gut microbiota in patients with asthenozoospermia: a Chinese pilot study
BMC Microbiology (2024)
-
Altered oral microbiome, but normal human papilloma virus prevalence in cartilage-hair hypoplasia patients
Orphanet Journal of Rare Diseases (2024)
-
The constitutive activation of STAT3 gene and its mutations are at the crossroad between LGL leukemia and autoimmune disorders
Blood Cancer Journal (2024)
-
The gut microbiome in systemic lupus erythematosus: lessons from rheumatic fever
Nature Reviews Rheumatology (2024)
-
Balancing Act of the Intestinal Antimicrobial Proteins on Gut Microbiota and Health
Journal of Microbiology (2024)