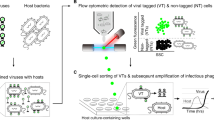
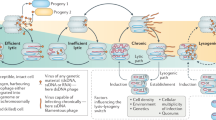
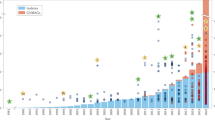
Abstract
Viruses are extremely diverse and modulate important biological and ecological processes globally. However, much of viral diversity remains uncultured and yet to be discovered. Several powerful culture-independent tools, in particular metagenomics, have substantially advanced virus discovery. Among those tools is single-virus genomics, which yields sequenced reference genomes from individual sorted virus particles without the need for cultivation. This new method complements virus culturing and metagenomic approaches and its advantages include targeted investigation of specific virus groups and investigation of genomic microdiversity within viral populations. In this Review, we provide a brief history of single-virus genomics, outline how this emergent method has facilitated advances in virus ecology and discuss its current limitations and future potential. Finally, we address how this method may synergistically intersect with other single-virus and single-cell approaches.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Koonin, E. V. The wonder world of microbial viruses. Expert Rev. Anti Infect. Ther. 8, 1097–1099 (2010).
Yong, E. I Contain Multitudes: The Microbes Within Us and A Grander View of Life (Ecco, 2016).
Breitbart, M. & Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284 (2005).
Paez-Espino, D. et al. Uncovering Earth’s virome. Nature 536, 425–430 (2016). This is a massive metagenomic study on global viral diversity and distribution and host specificity of viruses. A total of 125,000 partial DNA virus genomes are discovered.
Edwards, R. A. & Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 3, 504–510 (2005).
Suttle, C. A. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007). This is a fundamental must-read review of the general role of viruses in marine ecosystems.
Abedon, S. T. Bacteriophage Ecology: Population Growth, Evolution, and Impact of Bacterial Viruses (Cambridge Univ. Press, 2008).
Sullivan, M. B., Waterbury, J. B. & Chisholm, S. W. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424, 1047–1051 (2003).
Sullivan, M. B. et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12, 3035–3056 (2010).
Kauffman, K. M. et al. A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554, 118–122 (2018).
Atanasova, N. S., Roine, E., Oren, A., Bamford, D. H. & Oksanen, H. M. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 14, 426–440 (2012).
Marston, M. F. et al. Rapid diversification of coevolving marine Synechococcus and a virus. Proc. Natl Acad. Sci. USA 109, 4544–4549 (2012).
Enav, H., Kirzner, S., Lindell, D., Mandel-Gutfreund, Y. & Béjà, O. Adaptation to sub-optimal hosts is a driver of viral diversification in the ocean. Nat. Commun. 9, 1–11 (2018).
Rappé, M. S. & Giovannoni, S. J. The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003). This is a comprehensive review addressing a fundamental question in microbial ecology on the difficulty of culturing most microorganisms in the laboratory and how this bias impacts microbial discovery.
Pedrós-Alió, C. The rare bacterial biosphere. Ann. Rev. Mar. Sci. 4, 449–466 (2012).
Brum, J. R., Schenck, R. O. & Sullivan, M. B. Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses. ISME J. 7, 1738–1751 (2013).
Brum, J. R. et al. Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015). This is a pioneering, comprehensive metagenomic study on global marine viral diversity from hundreds of samples collected during the Tara expedition.
Trubl, G. et al. Soil viruses are underexplored players in ecosystem carbon processing. mSystems 3, e00076-18 (2018).
Paez-Espino, D. et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 47, D678–D686 (2019). This article describes the most comprehensive genome database of uncultured viruses recovered by metagenomics from different ecosystems, including the human body, with more than 700,000 viral genome fragments.
Carlson, C. J., Zipfel, C. M., Garnier, R. & Bansal, S. Global estimates of mammalian viral diversity accounting for host sharing. Nat. Ecol. Evol. 3, 1070–1075 (2019).
Carroll, D. et al. The Global Virome Project. Science 359, 872–874 (2018).
Cesar Ignacio-Espinoza, J., Solonenko, S. A. & Sullivan, M. B. The global virome: not as big as we thought? Curr. Opin. Virol. 3, 566–571 (2013). The authors address a hot topic in viral ecology (that is, how big the viral diversity in nature is) and estimate the total number of different viral proteins, which is a proxy for quantifying the number of different existing viruses.
Rohwer, F. Global phage diversity. Cell 113, 141 (2003).
Suttle, C. A. Environmental microbiology: viral diversity on the global stage. Nat. Microbiol. 1, 1–2 (2016).
Roux, S. et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693 (2016).
Schulz, F. et al. Hidden diversity of soil giant viruses. Nat. Commun. 9, 1–9 (2018). The article reports the discovery of several relevant giant viruses, including one with a genome of 2.4 Mb, using metagenomics and a method that is similar to those used in SVG, but in this case targeting multiple sets of 100 viruses, instead of single-virus particles.
Brown, C. T. et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211 (2015).
Anantharaman, K. et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 1–11 (2016).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Schulz, F. et al. Giant virus diversity and host interactions through global metagenomics. Nature 578, 432–436 (2020).
Al-Shayeb, B. et al. Clades of huge phages from across Earth’s ecosystems. Nature 578, 425–431 (2020).
Breitbart, M. et al. Genomic analysis of uncultured marine viral communities. Proc. Natl Acad. Sci. USA 99, 14250–14255 (2002).
Dávila-Ramos, S. et al. A review on viral metagenomics in extreme environments. Front. Microbiol. 10, 2403 (2019).
Chatterjee, A., Sicheritz-Pontén, T., Yadav, R. & Kondabagil, K. Genomic and metagenomic signatures of giant viruses are ubiquitous in water samples from sewage, inland lake, waste water treatment plant, and municipal water supply in Mumbai, India. Sci. Rep. 9, 1–9 (2019).
Simmonds, P. et al. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 15, 161–168 (2017).
Martinez-Hernandez, F. et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat. Commun. 8, 1–13 (2017). This is a pioneering reference high-throughput SVG study that unveils extremely abundant and ubiquitous uncultured marine viruses overlooked for years by current state-of-the-art, standard metagenomic-based studies.
Roux, S., Emerson, J. B., Eloe-Fadrosh, E. A. & Sullivan, M. B. Benchmarking viromics: an in silico evaluation of metagenome-enabled estimates of viral community composition and diversity. PeerJ 5, e3817 (2017). This in silico study performs a through bioinformatic comparison of different tools used commonly in viral metagenomics and aims to provide useful recommendations and standards for the scientific community.
Aguirre de Cárcer, D., Angly, F. E. & Alcamí, A. Evaluation of viral genome assembly and diversity estimation in deep metagenomes. BMC Genomics 15, 1–12 (2014).
López-Pérez, M., Haro-Moreno, J. M., Gonzalez-Serrano, R., Parras-Moltó, M. & Rodriguez-Valera, F. Genome diversity of marine phages recovered from Mediterranean metagenomes: size matters. PLoS Genet. 13, e1007018 (2017).
Labonté, J. M. et al. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J. 9, 2386–2399 (2015). The screening of sequencing data from hundreds of single cells obtained from seawater unveils virus–host interactions in different ecologically important bacterial and archaeal groups.
Roux, S. et al. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLife 2014, e03125 (2014).
Yoon, H. S. et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332, 714–717 (2011). This is the first report of SCG in uncultivated widespread microbial eukaryotes, showing complex viral interactions and metabolic insights into phycobiliphyte groups.
Castillo, Y. M. et al. Assessing the viral content of uncultured picoeukaryotes in the global‐ocean by single cell genomics. Mol. Ecol. 28, 4272–4289 (2019).
Benites, L. F. et al. Single cell ecogenomics reveals mating types of individual cells and ssDNA viral infections in the smallest photosynthetic eukaryotes. Phil. Trans. R. Soc. B 374, 20190089 (2019).
Martinez-Hernandez, F. et al. Single-cell genomics uncover Pelagibacter as the putative host of the extremely abundant uncultured 37-F6 viral population in the ocean. ISME J. 13, 232–236 (2019).
Brussaard, C. P. D., Noordeloos, A. A. M., Sandaa, R. A., Heldal, M. & Bratbak, G. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 319, 280–291 (2004).
Dean, F. B. et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA 99, 5261–5266 (2002).
Raghunathan, A. et al. Genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 71, 3342–3347 (2005).
Stepanauskas, R. & Sieracki, M. E. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc. Natl Acad. Sci. USA 104, 9052–9057 (2007).
Rinke, C. et al. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat. Protoc. 9, 1038–1048 (2014).
Martinez-Garcia, M., Martinez-Hernandez, F. & Martínez Martínez, J. Single-virus genomics: studying uncultured viruses, one at a time. Ref. Module Life Sci. https://doi.org/10.1016/b978-0-12-809633-8.21497-0 (2020). The authors provide methodological details and protocols for implementing SVG to complement other existing methods in viral ecology.
Lindell, D. et al. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl Acad. Sci. USA 101, 11013–11018 (2004).
Breitbart, M., Thompson, L., Suttle, C. & Sullivan, M. Exploring the vast diversity of marine viruses. Oceanography 20, 135–139 (2007).
Brum, J. R. & Sullivan, M. B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 13, 147–159 (2015). This review is recommended for readers who would like an introduction to recent technological advances in marine virology.
De Corte, D. et al. Viral communities in the global deep ocean conveyor belt assessed by targeted viromics. Front. Microbiol. 10, 1801 (2019).
Aylward, F. O. et al. Diel cycling and long-term persistence of viruses in the ocean’s euphotic zone. Proc. Natl Acad. Sci. USA 114, 11446–11451 (2017).
Luo, E., Aylward, F. O., Mende, D. R. & Delong, E. F. Bacteriophage distributions and temporal variability in the ocean’s interior. mBio 8, e01903-17 (2017).
Angly, F. E. et al. The marine viromes of four oceanic regions. PLoS Biol. 4, 2121–2131 (2006).
Coutinho, F. H., Rosselli, R. & Rodríguez-Valera, F. Trends of microdiversity reveal depth-dependent evolutionary strategies of viruses in the Mediterranean. mSystems 4, 1–17 (2019).
Roux, S., Krupovic, M., Debroas, D., Forterre, P. & Enault, F. Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open Biol. 3, 130160 (2013).
Zolfo, M. et al. Detecting contamination in viromes using ViromeQC. Nat. Biotechnol. 37, 1408–1412 (2019).
Amgarten, D., Braga, L. P. P., da Silva, A. M. & Setubal, J. C. MARVEL, a tool for prediction of bacteriophage sequences in metagenomic bins. Front. Genet. 9, 304 (2018).
Roux, S., Enault, F., Hurwitz, B. L. & Sullivan, M. B. VirSorter: mining viral signal from microbial genomic data. PeerJ 3, e985 (2015).
Ponsero, A. J. & Hurwitz, B. L. The promises and pitfalls of machine learning for detecting viruses in aquatic metagenomes. Front. Microbiol. 10, 806 (2019).
Crummett, L. T., Puxty, R. J., Weihe, C., Marston, M. F. & Martiny, J. B. H. The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 499, 219–229 (2016).
Pagarete, A., Allen, M. J., Wilson, W. H., Kimmance, S. A. & de Vargas, C. Host-virus shift of the sphingolipid pathway along an Emiliania huxleyi bloom: survival of the fattest. Environ. Microbiol. 11, 2840–2848 (2009).
Gregory, A. C. et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell 177, 1109–1123 (2019).
Kavagutti, V. S., Andrei, A. Ş., Mehrshad, M., Salcher, M. M. & Ghai, R. Phage-centric ecological interactions in aquatic ecosystems revealed through ultra-deep metagenomics. Microbiome 7, 1–15 (2019).
Sutton, T. D. S., Clooney, A. G., Ryan, F. J., Ross, R. P. & Hill, C. Choice of assembly software has a critical impact on virome characterisation. Microbiome 7, 12 (2019).
Madoui, M.-A. et al. Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genomics 16, 327 (2015).
Warwick-Dugdale, J. et al. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ 7, e6800 (2019). This pioneering study successfully combines long-read and short-read sequencing data to improve viral metagenomic assemblies and shows the potential of Nanopore sequencing data to advance virus discovery.
Beaulaurier, J. et al. Assembly-free single-molecule sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 30, 437–446 (2020).
Mizuno, C. M., Rodriguez-Valera, F., Kimes, N. E. & Ghai, R. Expanding the marine virosphere using metagenomics. PLoS Genet. 9, e1003987 (2013).
Garcia-Heredia, I. et al. Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. PLoS ONE 7, e33802 (2012).
Chow, C. E. T., Winget, D. M., White, R. A., Hallam, S. J. & Suttle, C. A. Combining genomic sequencing methods to explore viral diversity and reveal potential virus-host interactions. Front. Microbiol. 6, 265 (2015).
Mizuno, C. M., Ghai, R., Saghaï, A., López-García, P. & Rodriguez-Valera, F. Genomes of abundant and widespread viruses from the deep ocean. mBio 7, e00805–e00816 (2016).
Martinez-Garcia, M. et al. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 6, 113–123 (2012).
Stepanauskas, R. Single cell genomics: an individual look at microbes. Curr. Opin. Microbiol. 15, 613–620 (2012).
Sieracki, M. E. et al. Single cell genomics yields a wide diversity of small planktonic protists across major ocean ecosystems. Sci. Rep. 9, 1–11 (2019).
Lasken, R. S. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 10, 631–640 (2012).
López-Escardó, D. et al. Evaluation of single-cell genomics to address evolutionary questions using three SAGs of the choanoflagellate Monosiga brevicollis. Sci. Rep. 7, 1–14 (2017).
Mangot, J. F. et al. Accessing the genomic information of unculturable oceanic picoeukaryotes by combining multiple single cells. Sci. Rep. 7, 1–12 (2017).
Seeleuthner, Y. et al. Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nat. Commun. 9, 1–10 (2018).
Rinke, C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). This article is an excellent example of the power of single-cell technologies to provide biological insights into uncultured microorganisms.
Swan, B. K. et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc. Natl Acad. Sci. USA 110, 11463–11468 (2013).
Garcia, S. L. et al. Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J. 7, 137–147 (2013).
Martinez-Garcia, M. et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of verrucomicrobia. PLoS ONE 7, e35314 (2012).
Stepanauskas, R. et al. Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat. Commun. 8, 1–10 (2017). The authors use flow cytometry to sort uncultured single viruses and they amplify their genomes with a new variant of an efficient Φ29 enzyme, which is commonly used in SCG and SVG. This study is another SVG example targeting uncultured viruses.
Ghylin, T. W. et al. Comparative single-cell genomics reveals potential ecological niches for the freshwater acI Actinobacteria lineage. ISME J. 8, 2503–2516 (2014).
Wilson, W. H. et al. Genomic exploration of individual giant ocean viruses. ISME J. 11, 1736–1745 (2017). This reference SVG study targets for the first time uncultured giant viruses in nature, which are commonly ignored with standard metagenomic techniques.
de la Cruz Peña, M. et al. Deciphering the human virome with single-virus genomics and metagenomics. Viruses 10, 113 (2018). This is the first study on SVG applied to the human virome. The authors implement this novel technology, combined with metagenomics, in salivary human samples and discover important, abundant phages.
Allen, L. Z. et al. Single virus genomics: a new tool for virus discovery. PLoS ONE 6, e17722 (2011). This is the first report showing the feasibility of SVG as a new tool for virus discovery. The authors successfully use this technology to sequence several single sorted virus particles of viral isolates T4 and λ of E. coli.
Holmfeldt, K., Odić, D., Sullivan, M. B., Middelboe, M. & Riemann, L. Cultivated single-stranded DNA phages that infect marine bacteroidetes prove difficult to detect with DNA-binding stains. Appl. Environ. Microbiol. 78, 892–894 (2012).
Pospichalova, V. et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 4, 25530 (2015).
Giesecke, C. et al. Determination of background, signal-to-noise, and dynamic range of a flow cytometer: a novel practical method for instrument characterization and standardization. Cytometry A 91, 1104–1114 (2017).
Schmidt, H. & Hawkins, A. R. Single-virus analysis through chip-based optical detection. Bioanalysis 8, 867–870 (2016).
Brussaard, C., Payet, J. P., Winter, C. & Weinbauer, M. G. Quantification of aquatic viruses by flow cytometry. Man. Aquat. Viral Ecol. 11, 102–109 (2010).
Mojica, K. D. A. & Brussaard, C. P. D. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 89, 495–515 (2014).
Blainey, P. C. & Quake, S. R. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 39, e19 (2011).
Woyke, T. et al. Decontamination of MDA reagents for single cell whole genome amplification. PLoS ONE 6, e26161 (2011).
Povilaitis, T., Alzbutas, G., Sukackaite, R., Siurkus, J. & Skirgaila, R. In vitro evolution of phi29 DNA polymerase using isothermal compartmentalized self replication technique. Protein Eng. Des. Sel. 29, 617–628 (2016).
Gawad, C., Koh, W. & Quake, S. R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016). This is one of the most comprehensive technical and scientific reviews of SCG technologies of unicellular and multicellular organisms, and discusses how these technologies have enabled new discoveries in multiple fields from microbiology to cancer or immunology.
Martínez Martínez, J., Swan, B. K. & Wilson, W. H. Marine viruses, a genetic reservoir revealed by targeted viromics. ISME J. 8, 1079–1088 (2014). This study uses technologies similar to those used in SVG to discover giant viruses and other relevant uncultured viruses from a sorted pool of marine uncultured viruses.
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Woyke, T. et al. One bacterial cell, one complete genome. PLoS ONE 5, e10314 (2010).
Roux, S. et al. Minimum information about an uncultivated virus genome (MIUVIG). Nat. Biotechnol. 37, 29–37 (2019).
Hercher, M., Mueller, W. & Shapiro, H. M. Detection and discrimination of individual viruses by flow cytometry. J. Histochem. Cytochem. 27, 350–352 (1979).
Lippé, R. Flow virometry: a powerful tool to functionally characterize viruses. J. Virol. 92, e01765-17 (2017).
Koonin, E. V. & Yutin, N. Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv. Virus Res. 103, 167–202 (2019).
Brum, J. R. et al. Illuminating structural proteins in viral ‘dark matter’ with metaproteomics. Proc. Natl Acad. Sci. USA 113, 2436–2441 (2016).
Alonso-Sáez, L., Morán, X. A. G. & Clokie, M. R. Low activity of lytic pelagiphages in coastal marine waters. ISME J. 12, 2100–2102 (2018).
Zhao, Y. et al. Abundant SAR11 viruses in the ocean. Nature 494, 357–360 (2013).
McMullen, A., Martinez‐Hernandez, F. & Martinez‐Garcia, M. Absolute quantification of infecting viral particles by chip‐based digital polymerase chain reaction. Environ. Microbiol. Rep. 11, 855–860 (2019).
Fukuda, R., Ogawa, H., Nagata, T. & Koike, I. Direct determination of carbon and nitrogen contents of natural bacterial assemblages in marine environments. Appl. Environ. Microbiol. 64, 3352–3358 (1998).
Needham, D. M. et al. Targeted metagenomic recovery of four divergent viruses reveals shared and distinctive characteristics of giant viruses of marine eukaryotes. Phil. Trans. R. Soc. B 374, 20190086 (2019).
Needham, D. M. et al. A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc. Natl Acad. Sci. USA 116, 20574–20583 (2019).
Dieterich, D. C., Link, A. J., Graumann, J., Tirrell, D. A. & Schuman, E. M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl Acad. Sci. USA 103, 9482–9487 (2006).
Hatzenpichler, R. et al. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ. Microbiol. 16, 2568–2590 (2014).
Pasulka, A. L. et al. Interrogating marine virus-host interactions and elemental transfer with BONCAT and nanoSIMS-based methods. Environ. Microbiol. 20, 671–692 (2018).
Dominguez-Medina, S. et al. Neutral mass spectrometry of virus capsids above 100 megadaltons with nanomechanical resonators. Science 362, 918–922 (2018).
Hermelink, A. et al. Towards a correlative approach for characterising single virus particles by transmission electron microscopy and nanoscale Raman spectroscopy. Analyst 142, 1342–1349 (2017).
Ruokola, P. et al. Raman spectroscopic signatures of echovirus 1 uncoating. J. Virol. 88, 8504–8513 (2014).
Schatz, D. et al. Communication via extracellular vesicles enhances viral infection of a cosmopolitan alga. Nat. Microbiol. 2, 1485–1492 (2017).
Berleman, J. & Auer, M. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ. Microbiol. 15, 347–354 (2013).
Van Niel, G., D’Angelo, G. & Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018).
Machtinger, R., Laurent, L. C. & Baccarelli, A. A. Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum. Reprod. Update 22, 182–193 (2016).
Biller, S. J. et al. Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates. ISME J. 11, 394–404 (2017).
Kulp, A. & Kuehn, M. J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184 (2010).
Jacob, F. & Wollman, E. L. Viruses and genes. Sci. Am. 204, 93–107 (1961).
Forterre, P. The virocell concept and environmental microbiology. ISME J. 7, 233–236 (2013).
Forterre, P. Manipulation of cellular syntheses and the nature of viruses: the virocell concept. C. R. Chim. 14, 392–399 (2011).
Weitz, J. S., Li, G., Gulbudak, H., Cortez, M. H. & Whitaker, R. J. Viral invasion fitness across a continuum from lysis to latency. Virus Evol. 5, vez006 (2019).
Martinez-Garcia, M. et al. Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J. 6, 703–707 (2012).
Martínez-García, M., Santos, F., Moreno-Paz, M., Parro, V. & Antón, J. Unveiling viral–host interactions within the ‘microbial dark matter’. Nat. Commun. 5, 1–8 (2014).
Džunková, M. et al. Defining the human gut host–phage network through single-cell viral tagging. Nat. Microbiol. 4, 2192–2203 (2019). This is probably one of the most comprehensive SCG studies within the context of the human gut microbiota, and unveils a total of 363 unique host–phage pairings, expanding the known host–phage network of the gut microbiota.
Munson-Mcgee, J. H. et al. A virus or more in (nearly) every cell: Ubiquitous networks of virus-host interactions in extreme environments. ISME J. 12, 1706–1714 (2018).
Jarett, J. K. et al. Insights into the dynamics between viruses and their hosts in a hot spring microbial mat. ISME J. 14, 2527–2541 (2020).
Deng, L. et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513, 242–245 (2014).
Allers, E. et al. Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ. Microbiol. 15, 2306–2318 (2013).
Zanini, F. et al. Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue. Proc. Natl Acad. Sci. USA 115, E12363–E12369 (2018).
Steuerman, Y. et al. Dissection of influenza infection in vivo by single-cell RNA sequencing. Cell Syst. 6, 679–691 (2018).
Wyler, E. et al. Single-cell RNA-sequencing of herpes simplex virus 1-infected cells connects NRF2 activation to an antiviral program. Nat. Commun. 10, 1–14 (2019).
Guo, Q., Duffy, S. P., Matthews, K., Islamzada, E. & Ma, H. Deformability based cell sorting using microfluidic ratchets enabling phenotypic separation of leukocytes directly from whole blood. Sci. Rep. 7, 1–11 (2017).
Liu, W. et al. More than efficacy revealed by single-cell analysis of antiviral therapeutics. Sci. Adv. 5, eaax4761 (2019).
Lasken, R. S. Single-cell genomic sequencing using multiple displacement amplification. Curr. Opin. Microbiol. 10, 510–516 (2007).
Marcy, Y. et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl Acad. Sci. USA 104, 11889–11894 (2007).
Swan, B. K. et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1300 (2011).
Ahrendt, S. R. et al. Leveraging single-cell genomics to expand the fungal tree of life. Nat. Microbiol. 3, 1417–1428 (2018).
McConnell, M. J. et al. Mosaic copy number variation in human neurons. Science 342, 632–637 (2013).
Poulin, J. F., Tasic, B., Hjerling-Leffler, J., Trimarchi, J. M. & Awatramani, R. Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141 (2016).
Tanay, A. & Regev, A. Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338 (2017).
Sandberg, R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods 11, 22–24 (2014).
Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014).
Lindell, D. et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449, 83–86 (2007).
Roux, S., Tournayre, J., Mahul, A., Debroas, D. & Enault, F. Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinformatics 15, 1–12 (2014).
Watson, M., Schnettler, E. & Kohl, A. viRome: an R package for the visualization and analysis of viral small RNA sequence datasets. Bioinformatics 29, 1902–1903 (2013).
Jurtz, V. I., Villarroel, J., Lund, O., Voldby Larsen, M. & Nielsen, M. MetaPhinder—identifying bacteriophage sequences in metagenomic data sets. PLoS ONE 11, e0163111 (2016).
Zheng, T. et al. Mining, analyzing, and integrating viral signals from metagenomic data. Microbiome 7, 1–15 (2019).
Ren, J., Ahlgren, N. A., Lu, Y. Y., Fuhrman, J. A. & Sun, F. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 5, 69 (2017).
Fang, Z. et al. PPR-Meta: a tool for identifying phages and plasmids from metagenomic fragments using deep learning. GigaScience 8, giz066 (2019).
Tampuu, A., Bzhalava, Z., Dillner, J. & Vicente, R. ViraMiner: Deep learning on raw DNA sequences for identifying viral genomes in human samples. PLoS ONE 14, e0222271 (2019).
Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019).
Schleyer, G. et al. In plaque-mass spectrometry imaging of a bloom-forming alga during viral infection reveals a metabolic shift towards odd-chain fatty acid lipids. Nat. Microbiol. 4, 527–538 (2019).
Van Etten, J. L., Burbank, D. E., Kuczmarski, D. & Meints, R. H. Virus infection of culturable Chlorella-like algae and development of a plaque assay. Science 219, 994–996 (1983).
Maxwell, K. L. & Frappier, L. Viral proteomics. Microbiol. Mol. Biol. Rev. 71, 398–411 (2007).
Lum, K. K. & Cristea, I. M. Proteomic approaches to uncovering virus-host protein interactions during the progression of viral infection. Expert Rev. Proteom. 13, 325–340 (2016).
Cheng, W. & Schimert, K. A method for tethering single viral particles for virus-cell interaction studies with optical tweezers. Proc. SPIE 10723, 107233B (2018).
Ekeberg, T. et al. Three-dimensional reconstruction of the giant mimivirus particle with an X-ray free-electron laser. Phys. Rev. Lett. 114, 098102 (2015).
Cheng, Y. Single-particle cryo-EM at crystallographic resolution. Cell 161, 450–457 (2015).
Lyumkis, D. Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 294, 5181–5197 (2019).
Subramaniam, S., Bartesaghi, A., Liu, J., Bennett, A. E. & Sougrat, R. Electron tomography of viruses. Curr. Opin. Struct. Biol. 17, 596–602 (2007).
Gamage, S. et al. Probing structural changes in single enveloped virus particles using nano-infrared spectroscopic imaging. PLoS ONE 13, e0199112 (2018).
Martínez Martínez, J., Schroeder, D. C., Larsen, A., Bratbak, G. & Wilson, W. H. Molecular dynamics of Emiliania huxleyi and cooccurring viruses during two separate mesocosm studies. Appl. Environ. Microbiol. 73, 554–562 (2007).
Martínez Martínez, J. et al. New lipid envelope-containing dsDNA virus isolates infecting Micromonas pusilla reveal a separate phylogenetic group. Aquat. Microb. Ecol. 74, 17–28 (2015).
Acknowledgements
This work was supported by the Gordon and Betty Moore Foundation (grant 5334), the US National Science Foundation (NSF-OPP 1644155, NSF-OCE 1933289), the Spanish Ministry of Economy and Competitiveness (CGL2013-40564-R, RTI2018-094248-B-I00 and SAF2013-49267-EXP) and Generalitat Valenciana (ACOM/2015/133 and ACIF/2015/332).
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, contributed to the discussion of the content and reviewed and edited the manuscript before submission. M.M.-G. and J.M.M. wrote the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks Julio Cesar Ignacio Espinoza and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- IMG/VR database
-
Integrated data management and analysis system for cultivated and environmental viral genomes, which is publicly available for the scientific community.
- Metagenomics
-
The study of sequenced nucleic acids obtained from bulk environmental samples (enriched in cells or viruses).
- Tara expedition
-
Oceanic 3-year-long expedition around the world to investigate planktonic and coral ecosystems and the impacts of global change. More than 150 international scientists have taken part.
- Auxiliary metabolic genes
-
Cellular host genes contained in the viral genome that modulate cellular metabolism during infection to improve viral replication.
- Nucleocytoplasmic large DNA viruses
-
(NCLDVs). A group of large DNA viruses with genomes ranging from 150 kb to 1.2 Mb classified within the phylum Nucleocytoviricota. These viruses are referred to as ‘nucleocytoplasmic’ because they are often able to replicate in both the host cell nucleus and the host cell cytoplasm.
- Contigs
-
High-confidence overlapped DNA sequenced reads that represent a consensus region of a genome.
- Flow cytometry
-
Technique used to detect and measure some physical and chemical features of a population of cells, viruses or particles suspended in a fluid that flow one at a time through a laser beam, where the light scattered is detected along with other fluorescence features. The sample is often fluorescently stained with cell and/or virus markers.
- Single amplified genomes
-
(SAGs). Genome sequences obtained from sequencing and assembly of the amplified genetic material from an individual sorted single cell.
- Multiple-displacement amplification
-
Common whole-genome amplification technique used in single-cell genomics to amplify minute amounts of DNA. DNA synthesis and amplification is done by Φ29 DNA polymerase.
- Virions
-
Complete virus particles, in their extracellular phase, that are able to carry out the infectious process. Typically, the viral genome is enclosed in a protein structure (capsid) and is sometimes surrounded by a lipid membrane.
- Gene-transfer agents
-
Phage-like entities that contain only a random piece of the cellular genome, which is insufficient to encode its protein components.
- Consensus sequences
-
The calculated order of the most frequent residues, either nucleotide or amino acid, found at each position in a sequence.
- Ultradeep sequencing
-
DNA sequencing performed at very high coverage. ‘Deep sequencing’ refers to sequencing a genomic region multiple times, sometimes hundreds or even thousands of times.
- Fosmids
-
Clone system based on the bacterial F plasmid usually in Escherichia coli that can hold a DNA insert of up to 40 kb in size.
- Deep chlorophyll maximum
-
Region below the surface of water with the maximum concentration of chlorophyll.
- Viral single amplified genomes
-
(vSAGs). Genome sequences obtained from sequencing and assembly of the amplified genetic material from an individual sorted single-virus particle.
- Viral shunt
-
Mechanism mediated by viral infection and consequently cell lysis that prevents (prokaryotic and eukaryotic) marine microbial particulate organic matter from migrating up trophic levels by recycling it into dissolved organic matter.
Rights and permissions
About this article
Cite this article
Martínez Martínez, J., Martinez-Hernandez, F. & Martinez-Garcia, M. Single-virus genomics and beyond. Nat Rev Microbiol 18, 705–716 (2020). https://doi.org/10.1038/s41579-020-00444-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-020-00444-0
This article is cited by
-
Viruses under the Antarctic Ice Shelf are active and potentially involved in global nutrient cycles
Nature Communications (2023)
-
Genomics discovery of giant fungal viruses from subsurface oceanic crustal fluids
ISME Communications (2023)
-
Giant virus biology and diversity in the era of genome-resolved metagenomics
Nature Reviews Microbiology (2022)
-
Unexpected myriad of co-occurring viral strains and species in one of the most abundant and microdiverse viruses on Earth
The ISME Journal (2022)