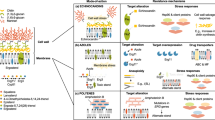
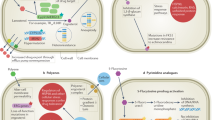
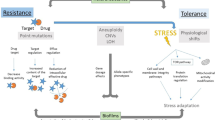
Abstract
Systemic fungal infections pose a serious clinical problem. Treatment options are limited, and antifungal drug resistance is increasing. In addition, a substantial proportion of patients do not respond to therapy despite being infected with fungi that are susceptible to the drug. The discordance between overall treatment outcome and low levels of clinical resistance may be attributable to antifungal drug tolerance. In this Review, we define and distinguish resistance and tolerance and discuss the current understanding of the molecular, genetic and physiological mechanisms that contribute to those phenomena. Distinguishing tolerance from resistance might provide important insights into the reasons for treatment failure in some settings.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Arendrup, M. C. & Patterson, T. F. Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216, S445–S451 (2017).
Perlin, D. S., Rautemaa-Richardson, R. & Alastruey-Izquierdo, A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect. Dis. 17, e383–e392 (2017).
Alangaden, G. J. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect. Dis. Clin. North Am. 25, 201–225 (2011).
Andes, D. R. et al. The epidemiology and outcomes of invasive Candida infections among organ transplant recipients in the United States: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Transpl. Infect. Dis. 18, 921–931 (2016).
Krysan, D. J. The unmet clinical need of novel antifungal drugs. Virulence 8, 135–137 (2017).
Bassetti, M., Carnelutti, A., Castaldo, N. & Peghin, M. Important new therapies for methicillin-resistant Staphylococcus aureus. Expert Opin. Pharmacother. 20, 2317–2334 (2019).
Pfaller, M. A., Diekema, D. J., Turnidge, J. D., Castanheira, M. & Jones, R. N. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997–2016. Open Forum Infect. Dis. 6, S79–S94 (2019).
Lepak, A. J. & Andes, D. R. Antifungal pharmacokinetics and pharmacodynamics. Cold Spring Harb. Perspect. Med. 5, a019653 (2014).
Rosenberg, A. et al. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat. Commun. 9, 2470 (2018). This article provides a quantitative characterization of fluconazole tolerance in C. albicans, distinguishing it from resistance and highlighting its phenotypic heterogeneity and association with subpopulation growth, and sensitivity to adjuvant drugs.
Brauner, A., Fridman, O., Gefen, O. & Balaban, N. Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016).
Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019).
Brauner, A., Shoresh, N., Fridman, O. & Balaban, N. Q. An experimental framework for quantifying bacterial tolerance. Biophys. J. 112, 2664–2671 (2017).
Levin-Reisman, I., Fridman, O. & Balaban, N. Q. ScanLag: high-throughput quantification of colony growth and lag time. J. Vis. Exp. https://doi.org/10.3791/51456 (2014).
Wuyts, J., Van Dijck, P. & Holtappels, M. Fungal persister cells: the basis for recalcitrant infections? PLoS Pathog. 14, e1007301 (2018).
Astvad, K. M. T., Sanglard, D., Delarze, E., Hare, R. K. & Arendrup, M. C. Implications of the EUCAST trailing phenomenon in Candida tropicalis for the in vivo susceptibility in invertebrate and murine models. Antimicrob. Agents Chemother. 62, e01624–18 (2018).
Marcos-Zambrano, L. J., Escribano, P., Sanchez-Carrillo, C., Bouza, E. & Guinea, J. Scope and frequency of fluconazole trailing assessed using EUCAST in invasive Candida spp. isolates. Med. Mycol. 54, 733–739 (2016).
Coenye, T., De Vos, M., Vandenbosch, D. & Nelis, H. Factors influencing the trailing endpoint observed in Candida albicans susceptibility testing using the CLSI procedure. Clin. Microbiol. Infect. 14, 495–497 (2008).
Agrawal, D., Patterson, T. F., Rinaldi, M. G. & Revankar, S. G. Trailing end-point phenotype of Candida spp. in antifungal susceptibility testing to fluconazole is eliminated by altering incubation temperature. J. Med. Microbiol. 56, 1003–1004 (2007).
Marr, K. A., Rustad, T. R., Rex, J. H. & White, T. C. The trailing end point phenotype in antifungal susceptibility testing is pH dependent. Antimicrob. Agents Chemother. 43, 1383–1386 (1999).
Delarze, E. & Sanglard, D. Defining the frontiers between antifungal resistance, tolerance and the concept of persistence. Drug Resist. Updat. 23, 12–19 (2015).
Marchetti, O., Moreillon, P., Glauser, M. P., Bille, J. & Sanglard, D. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 44, 2373–2381 (2000).
Taff, H. T., Mitchell, K. F., Edward, J. A. & Andes, D. R. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 8, 1325–1337 (2013).
Pappas, P. G. et al. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 62, 409–417 (2016).
Meletiadis, J., Curfs-Breuker, I., Meis, J. F. & Mouton, J. W. In vitro antifungal susceptibility testing of Candida isolates with the EUCAST methodology, a new method for ECOFF determination. Antimicrob. Agents Chemother. 61, e02372–16 (2017).
Lockhart, S. R., Ghannoum, M. A. & Alexander, B. D. Establishment and use of epidemiological cutoff values for molds and yeasts by use of the Clinical and Laboratory Standards Institute M57 standard. J. Clin. Microbiol. 55, 1262–1268 (2017). This review provides an important and accessible over-review of the concept of epidemiological cut-off values as well as the methodology that underlies their establishment.
Onyewu, C., Wormley, F. L. Jr., Perfect, J. R. & Heitman, J. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infect. Immun. 72, 7330–7333 (2004).
Sanglard, D., Ischer, F., Marchetti, O., Entenza, J. & Bille, J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48, 959–976 (2003).
Luna-Tapia, A., Tournu, H., Peters, T. L. & Palmer, G. E. Endosomal trafficking defects can induce calcium-dependent azole tolerance in Candida albicans. Antimicrob. Agents Chemother. 60, 7170–7177 (2016).
Luna-Tapia, A., Butts, A. & Palmer, G. E. Loss of C-5 sterol desaturase activity in Candida albicans: azole resistance or merely trailing growth? Antimicrob. Agents Chemother. 63, e00129–11 (2019).
van der Linden, J. W. et al. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin. Infect. Dis. 57, 513–520 (2013).
McCarty, T. P. & Pappas, P. G. Invasive candidiasis. Infect. Dis. Clin. North Am. 30, 103–124 (2016).
Prasad, R., Rawal, M. K. & Shah, A. H. Candida efflux ATPases and antiporters in clinical drug resistance. Adv. Exp. Med. Biol. 892, 351–376 (2016).
Sasse, C. et al. The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol. Microbiol. 86, 539–556 (2012). This article shows that when a strain accumulates multiple resistance mutations that affect different transcription factors, the effects are independent and additive. In addition, these effects, generated by engineered deletion mutations, are associated with reduced fitness in the absence of drug stress. As many resistant isolates do not have obvious fitness defects, it is assumed that they must have acquired compensatory mechanisms in addition to the characterized mechanisms of drug resistance.
Holmes, A. R. et al. Heterozygosity and functional allelic variation in the Candida albicans efflux pump genes CDR1 and CDR2. Mol. Microbiol. 62, 170–186 (2006).
Munoz, J. F. et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 9, 5346 (2018).
Flowers, S. A. et al. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 11, 1289–1299 (2012). This study establishes that gain-of-function mutations in UPC2, the key regulator of ergosterol biosynthesis gene expression, are a source of clinically important fluconazole resistance. Previously, only transcriptional regulators of efflux pump expression were known to do so.
Xiang, M. J. et al. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 13, 386–393 (2013).
Flowers, S. A., Colon, B., Whaley, S. G., Schuler, M. A. & Rogers, P. D. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob. Agents Chemother. 59, 450–460 (2015).
Healey, K. R. et al. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother. 62, e01427–18 (2018).
Chowdhary, A. et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73, 891–899 (2018).
Lockhart, S. R. et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64, 134–140 (2017).
Satoh, K. et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53, 41–44 (2009).
Kordalewska, M. et al. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother. 62, e00238–18 (2018).
Selmecki, A., Forche, A. & Berman, J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313, 367–370 (2006).
Anderson, M. Z., Saha, A., Haseeb, A. & Bennett, R. J. A chromosome 4 trisomy contributes to increased fluconazole resistance in a clinical isolate of Candida albicans. Microbiology 163, 856–865 (2017).
Yang, F. et al. Tolerance to caspofungin in Candida albicans is associated with at least three distinctive mechanisms that govern expression of FKS genes and cell wall remodeling. Antimicrob. Agents Chemother. 61, e00071–17 (2017).
Yang, F., Kravets, A., Bethlendy, G., Welle, S. & Rustchenko, E. Chromosome 5 monosomy of Candida albicans controls susceptibility to various toxic agents, including major antifungals. Antimicrob. Agents Chemother. 57, 5026–5036 (2013).
Yang, F. et al. Aneuploidy enables cross-adaptation to unrelated drugs. Mol. Biol. Evol. 36, 1768–1782 (2019).
Todd, R. T., Wikoff, T. D., Forche, A. & Selmecki, A. Genome plasticity in Candida albicans is driven by long repeat sequences. eLife. 8, e45954 (2019).
Coste, A. et al. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172, 2139–2156 (2006).
Selmecki, A. M., Dulmage, K., Cowen, L. E., Anderson, J. B. & Berman, J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5, e1000705 (2009).
Mount, H. O. et al. Global analysis of genetic circuitry and adaptive mechanisms enabling resistance to the azole antifungal drugs. PLoS Genet. 14, e1007319 (2018). This article screens a library of C. albicans deletion strains and identifies two genes that affect azole responses, have roles in stress response pathways and functions that are are suppressed by the acquisition of aneuploid chromosomes. Whether the mutations are affecting resistance or tolerance is not clear, but activities of the identified genes (vacuolar retrograde trafficking and a cell wall and polarity GTPase activating protein) suggest that the mechanism is involved in processes that affect tolerance rather than in directly affecting resistance.
Todd, R. T., Forche, A. & Selmecki, A. Ploidy variation in fungi: polyploidy, aneuploidy, and genome evolution. Microbiol. Spectr. 5, https://doi.org/10.1128/microbiolspec.FUNK-0051-2016 (2017).
Bennett, R. J., Forche, A. & Berman, J. Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb. Perspect. Med. 4, a019604 (2014).
Ford, C. B. et al. The evolution of drug resistance in clinical isolates of Candida albicans. eLife 4, e00662 (2015).
White, T. C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41, 1482–1487 (1997). This article presents the first detailed study of the molecular mechanisms underlying the development of fluconazole resistance over time in patients. It is a landmark study because of its patient-based approach and because it yielded strains that are still under study today.
Cowen, L. E. et al. Population genomics of drug resistance in Candida albicans. Proc. Natl Acad. Sci. USA 99, 9284–9289 (2002).
Forche, A. et al. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6, e110 (2008).
Harrison, B. D. et al. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 12, e1001815 (2014).
Forche, A. et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio 2, e00129–11 (2011).
Ene, I. V. et al. Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen. Proc. Natl Acad. Sci. USA 115, E8688–E8697 (2018).
Forche, A. et al. Rapid phenotypic and genotypic diversification after exposure to the oral host niche in Candida albicans. Genetics 209, 725–741 (2018).
Forche, A., May, G. & Magee, P. T. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot. Cell 4, 156–165 (2005).
Forche, A., Magee, P. T., Selmecki, A., Berman, J. & May, G. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182, 799–811 (2009).
Perlin, D. S. Echinocandin resistance in Candida. Clin. Infect. Dis. 61, S612–S617 (2015).
Farmakiotis, D., Tarrand, J. J. & Kontoyiannis, D. P. Drug-resistant Candida glabrata infection in cancer patients. Emerg. Infect. Dis. 20, 1833–1840 (2014).
Garcia-Effron, G., Lee, S., Park, S., Cleary, J. D. & Perlin, D. S. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53, 3690–3699 (2009).
Cowen, L. E., Sanglard, D., Howard, S. J., Rogers, P. D. & Perlin, D. S. Mechanisms of antifungal drug resistance. CSH Perspect. Med. 5, a019752 (2015).
Bordallo-Cardona, M. A. et al. MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob. Agents Chemother. 63, e01876–18 (2019).
Singh, A. et al. Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob. Agents Chemother. 62, e00195–18 (2018).
Shor, E., Schuyler, J. & Perlin, D. S. A novel, drug resistance-independent, fluorescence-based approach to measure mutation rates in microbial pathogens. mBio 10, e00120–19 (2019).
Ropars, J. et al. Gene flow contributes to diversification of the major fungal pathogen Candida albicans. Nat. Commun. 9, 2253 (2018).
Hickman, M. A. et al. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494, 55–59 (2013).
Gao, J. et al. Candida albicans gains azole resistance by altering sphingolipid composition. Nat. Commun. 9, 4495 (2018). This study uses a haploid C. albicans isolate and a transposon mutagenesis system to identify mutants that are able to improve growth in very high fluconazole levels. However, the degree to which the mutants are resistant versus tolerant is not addressed, as drug responses did not include MIC assays. The study provides convincing data showing that alterations in the sphingolipid composition of membranes is associated with the ability to grow and divide in the presence of fluconazole.
Segal, E. S. et al. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. mBio 9, e02048–18 (2018).
Walker, L. A., Gow, N. A. & Munro, C. A. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 57, 146–154 (2013). This study finds that the ability of Candida species to grow in the presence of sub-MIC levels of caspofungin is associated with increased chitin levels in the cell wall. Pretreatment of cells with CaCl 2 and calcofluor white induces a similar increase in chitin and ability to grow in sub-MIC caspofungin levels. Whether this effect is an alteration in tolerance or resistance levels remains to be determined; higher cell wall chitin levels are induced by reagents, such as CaCl 2 and calcofluor white, and the effect is transient and physiological, thus sharing some features of tolerance.
Lee, K. K. et al. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 56, 208–217 (2012).
Singh-Babak, S. D. et al. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 8, e1002718 (2012).
Selmecki, A., Forche, A. & Berman, J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9, 991–1008 (2010).
Sitterle, E. et al. Within-host genomic diversity of Candida albicans in healthy carriers. Sci. Rep. 9, 2563 (2019).
Cowen, L. E. Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog. 5, e1000471 (2009).
Cowen, L. E. & Lindquist, S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309, 2185–2189 (2005).
Khandelwal, N. K. et al. Azole resistance in a Candida albicans mutant lacking the ABC transporter CDR6/ROA1 depends on TOR signaling. J. Biol. Chem. 293, 412–432 (2018).
Garnaud, C. et al. The Rim pathway mediates antifungal tolerance in Candida albicans through newly identified Rim101 transcriptional targets, including Hsp90 and Ipt1. Antimicrob. Agents Chemother. 62, e01785-17 (2018).
Robbins, N., Caplan, T. & Cowen, L. E. Molecular evolution of antifungal drug resistance. Annu. Rev. Microbiol. 71, 753–775 (2017).
Shapiro, R. S., Zaas, A. K., Betancourt-Quiroz, M., Perfect, J. R. & Cowen, L. E. The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PLoS One 7, e44734 (2012).
Cowen, L. E. et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl Acad. Sci. USA 106, 2818–2823 (2009).
Olin-Sandoval, V. et al. Lysine harvesting is an antioxidant strategy and triggers underground polyamine metabolism. Nature 572, 249–253 (2019).
Benhamou, R. I. et al. Real-time imaging of the azole class of antifungal drugs in live Candida cells. ACS Chem. Biol. 12, 1769–1777 (2017).
Benhamou, R. I., Bibi, M., Berman, J. & Fridman, M. Localizing antifungal drugs to the correct organelle can markedly enhance their efficacy. Angew. Chem. 57, 6230–6235 (2018).
Campbell, K., Herrera-Dominguez, L., Correia-Melo, C., Zelezniak, A. & Ralser, M. Biochemical principles enabling metabolic cooperativity and phenotypic heterogeneity at the single cell level. Curr. Opin. Syst. Biol. 8, 97–108 (2018).
Franzmann, T. M. & Alberti, S. Protein phase separation as a stress survival strategy. CSH Perspect. Biol. 11, a034058 (2019).
Delarue, M. et al. mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20 (2018).
Roemer, T. & Krysan, D. J. Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 4, a019703 (2014).
Cornet, M. et al. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect. Immun. 73, 7977–7987 (2005).
Gerstein, A. C., Rosenberg, A., Hecht, I. & Berman, J. diskImageR: quantification of resistance and tolerance to antimicrobial drugs using disk diffusion assays. Microbiology 162, 1059–1068 (2016).
Ben-Ami, R. et al. Heteroresistance to fluconazole is a continuously distributed phenotype among Candida glabrata clinical strains associated with in vivo persistence. mBio 7, e00655–16 (2016).
Sionov, E., Chang, Y. C., Garraffo, H. M. & Kwon-Chung, K. J. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob. Agents Chemother. 53, 2804–2815 (2009).
Varma, A. & Kwon-Chung, K. J. Heteroresistance of Cryptococcus gattii to fluconazole. Antimicrob. Agents Chemother. 54, 2303–2311 (2010).
Sionov, E., Chang, Y. C. & Kwon-Chung, K. J. Azole heteroresistance in Cryptococcus neoformans: emergence of resistant clones with chromosomal disomy in the mouse brain during fluconazole treatment. Antimicrob. Agents Chemother. 57, 5127–5130 (2013).
Stone, N. R. et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Invest. 129, 999–1014 (2019). This study tracks series of clinical Cryptococcus neoformans isolates from individual patients with HIV from Tanzania and finds that heteroresistance is quite common and is often, but not always, associated with specific chromosomal aneuploidies.
Desai, J. V., Mitchell, A. P. & Andes, D. R. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb. Perspect. Med. 4, a019729 (2014).
Lohse, M. B., Gulati, M., Johnson, A. D. & Nobile, C. J. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 16, 19–31 (2018).
Ramage, G., Vande Walle, K., Wickes, B. L. & Lopez-Ribot, J. L. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 45, 2475–2479 (2001).
Taff, H. T. et al. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 8, e1002848 (2012).
Li, P., Seneviratne, C. J., Alpi, E., Vizcaino, J. A. & Jin, L. Delicate metabolic control and coordinated stress response critically determine antifungal tolerance of Candida albicans biofilm persisters. Antimicrob. Agents Chemother. 59, 6101–6112 (2015).
Denega, I., d’Enfert, C. & Bachellier-Bassi, S. Candida albicans biofilms are generally devoid of persister cells. Antimicrob. Agents Chemother. 63, e01979–18 (2019).
Al-Dhaheri, R. S. & Douglas, L. J. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob. Agents Chemother. 52, 1884–1887 (2008).
Stevens, D. A., Espiritu, M. & Parmar, R. Paradoxical effect of caspofungin: reduced activity against Candida albicans at high drug concentrations. Antimicrob. Agents Chemother. 48, 3407–3411 (2004).
Stevens, D. A., White, T. C., Perlin, D. S. & Selitrennikoff, C. P. Studies of the paradoxical effect of caspofungin at high drug concentrations. Diagn. Microbiol. Infect. Dis. 51, 173–178 (2005).
Rueda, C., Cuenca-Estrella, M. & Zaragoza, O. Paradoxical growth of Candida albicans in the presence of caspofungin is associated with multiple cell wall rearrangements and decreased virulence. Antimicrob. Agents Chemother. 58, 1071–1083 (2014). This study presents a detailed assessment of the cellular changes that accompany growth beyond the MIC in caspofungin as well as the effects that those changes have on fitness in mammalian hosts.
Wagener, J. & Loiko, V. Recent insights into the paradoxical effect of echinocandins. J. Fungi 4, 5 (2017).
Rueda, C. et al. Evaluation of the possible influence of trailing and paradoxical effects on the clinical outcome of patients with candidemia. Clin. Microbiol. Infect. 23, 49.e1–49.e8 (2017).
Lee, M. K. et al. Susceptibility and trailing growth of Candida albicans to fluconazole: results of a Korean multicentre study. Mycoses 50, 148–149 (2007).
Arthington-Skaggs, B. A. et al. Comparison of visual and spectrophotometric methods of broth microdilution MIC end point determination and evaluation of a sterol quantitation method for in vitro susceptibility testing of fluconazole and itraconazole against trailing and nontrailing Candida isolates. Antimicrob. Agents Chemother. 46, 2477–2481 (2002).
Rex, J. H. et al. Optimizing the correlation between results of testing in vitro and therapeutic outcome in vivo for fluconazole by testing critical isolates in a murine model of invasive candidiasis. Antimicrob. Agents Chemother. 42, 129–134 (1998).
Colombo, A. L. et al. Prognostic factors and historical trends in the epidemiology of candidemia in critically ill patients: an analysis of five multicenter studies sequentially conducted over a 9-year period. Intensive Care Med. 40, 1489–1498 (2014).
Hirakawa, M. P. et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 25, 413–425 (2015).
Huang, M. Y., Woolford, C. A., May, G., McManus, C. J. & Mitchell, A. P. Circuit diversification in a biofilm regulatory network (vol 15, e1007787, 2019). PLoS Pathog. 15, e1007787 (2019).
Hooper, R. W., Ashcraft, D. S. & Pankey, G. A. In vitro synergy with fluconazole plus doxycycline or tigecycline against clinical Candida glabrata isolates. Med. Mycol. 57, 122–126 (2019).
Lu, M. et al. Linezolid in combination with azoles induced synergistic effects against Candida albicans and protected Galleria mellonella against experimental Candidiasis. Front. Microbiol. 9, 3142 (2018).
da Silva, C. R. et al. Synergistic effects of amiodarone and fluconazole on Candida tropicalis resistant to fluconazole. Antimicrob. Agents Chemother. 57, 1691–1700 (2013).
Li, H. et al. Resistance reversal induced by a combination of fluconazole and tacrolimus (FK506) in Candida glabrata. J. Med. Microbiol. 64, 44–52 (2015).
Gu, W., Guo, D., Zhang, L., Xu, D. & Sun, S. The synergistic effect of azoles and fluoxetine against resistant Candida albicans strains is attributed to attenuating fungal virulence. Antimicrob. Agents Chemother. 60, 6179–6188 (2016).
Costa-de-Oliveira, S. et al. Ibuprofen potentiates the in vivo antifungal activity of fluconazole against Candida albicans murine infection. Antimicrob. Agents Chemother. 59, 4289–4292 (2015).
Uppuluri, P., Nett, J., Heitman, J. & Andes, D. Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob. Agents Chemother. 52, 1127–1132 (2008).
Yu, S. J., Chang, Y. L. & Chen, Y. L. Calcineurin signaling: lessons from Candida species. FEMS Yeast Res. 15, fov016 (2015).
Hubbard, M., Bradley, M., Sullivan, P., Shepherd, M. & Forrester, I. Evidence for the occurrence of calmodulin in the yeasts Candida albicans and Saccharomyces cerevisiae. FEBS Lett. 137, 85–88 (1982).
O’Meara, T. R., Robbins, N. & Cowen, L. E. The Hsp90 chaperone network modulates Candida virulence traits. Trends Microbiol. 25, 809–819 (2017).
Acknowledgements
The authors thank M. Ralser, D. Jarosz and members of the Berman laboratory for helpful comments, and S. Everson, Iuliana Ene, Brown University, Anton Levitan, Tel-Aviv University, Aleeza C. Gerstein, University of Manitoba and M. Hajooj for help with illustrations. Work in the authors’ laboratories was supported by the European Research Council (RAPLODAPT 340087) and the Israel Science Foundation (grant number 997/18) (J.B.), and by the National Institutes of Health (1R01AI125094) (D.J.K.).
Author information
Authors and Affiliations
Contributions
Both authors contributed to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks J. Morschhauser, D. Perlin and D. Sanglard for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Candida Genome Database: http://www.candidagenome.org/
Supplementary information
Glossary
- Candidaemia
-
Candidaemia is a candidal infection of the blood stream.
- Susceptibility
-
Sensitivity to a drug, arresting growth (static drugs) and/or killing cells (cidal drugs).
- Fraction of growth
-
A measure of tolerance based on assays performed on a solid medium. Measured at 48 h, the growth within the zone of inhibition (and thus above the minimum inhibitory concentration) is estimated as a proportion of total growth possible outside the zone of inhibition.
- Supra-MIC growth
-
(SMG). A measure of tolerance based on assays performed in a liquid medium. Growth at concentrations above the minimum inhibitory concentration is estimated as a proportion of the total growth without a drug. SMG provides a quantitative measure of growth similar to some measures of trailing growth.
- Phenotypic heterogeneity
-
The expression of different phenotypes in different cells within an isogenic population of cells. For example, some fungal cells grow whereas other sister cells do not grow (or grow too slowly to be detected) in the presence of an antifungal drug.
- Fungistatic drugs
-
Drugs that inhibit growth but do not necessarily kill a majority of the cell population at concentrations at or above the minimum inhibitory concentration.
- Heteroresistance
-
A clinical term for isolates that contain small subpopulations of cells (generally <1%) that have the ability to grow at drug concentrations that are at least 8× the minimum inhibitory concentration for the vast majority of susceptible cells in the population.
- Trailing growth
-
Generally defined as reduced but persistent visible growth of Candida spp. at fluconazole concentrations above the minimum inhibitory concentration (MIC). Trailing has also been described as an increase in the MIC during growth beyond 24 h (the standard end point for MIC measurements for Candida spp.) and can be measured as the residual growth in the presence of fluconazole concentrations above the MIC. Trailing was quantified in a recent study as the percentage of residual yeast growth at fluconazole concentrations above the MIC in each well and mean trailing as the geometric mean of trailing observed in all of the wells above the MIC.
- Paradoxical growth
-
Also referred to as the Eagle effect. The ability of a fungal isolate to reconstitute growth in the presence of high drug concentrations, but being fully susceptible at lower concentrations. Paradoxical growth appears with a delay of one to several days, but resembles growth in the absence of the drug. Paradoxical growth has been reported primarily for echinocandins.
- Adjuvants
-
A drug that potentiates the effect of an anti-infective, but is not an anti-infective on its own.
- Fungicidal activity
-
Drugs with fungicidal activity reduce a population of cells by >99.9% or 3 log10 units at a concentration equal to or greater than the minimum inhibitory concentration.
Rights and permissions
About this article
Cite this article
Berman, J., Krysan, D.J. Drug resistance and tolerance in fungi. Nat Rev Microbiol 18, 319–331 (2020). https://doi.org/10.1038/s41579-019-0322-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-019-0322-2
This article is cited by
-
Metabolic sensing tips the balance of drug tolerance in fungal meningitis
Nature Microbiology (2024)
-
Brain glucose induces tolerance of Cryptococcus neoformans to amphotericin B during meningitis
Nature Microbiology (2024)
-
A dual-targeting antifungal is effective against multidrug-resistant human fungal pathogens
Nature Microbiology (2024)
-
Exploring the Potential of Farnesol as a Novel Antifungal Drug and Related Challenges
Current Infectious Disease Reports (2024)
-
Exploring the Therapeutic Potential of Scorpion-Derived Css54 Peptide Against Candida albicans
Journal of Microbiology (2024)