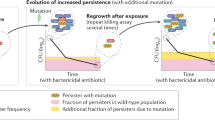
Abstract
Antibiotic heteroresistance is a phenotype in which a bacterial isolate contains subpopulations of cells that show a substantial reduction in antibiotic susceptibility compared with the main population. Recent work indicates that heteroresistance is very common for several different bacterial species and antibiotic classes. The resistance phenotype is often unstable, and in the absence of antibiotic pressure it rapidly reverts to susceptibility. A common mechanistic explanation for the instability is the occurrence of genetically unstable tandem amplifications of genes that cause resistance. Due to their instability, low frequency and transient character, it is challenging to detect and study these subpopulations, which often leads to difficulties in unambiguously classifying bacteria as susceptible or resistant. Finally, in vitro experiments, mathematical modelling, animal infection models and clinical studies show that the resistant subpopulations can be enriched during antibiotic exposure, and increasing evidence suggests that heteroresistance can lead to treatment failure.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hughes, D. & Andersson, D. I. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 41, 374–391 (2017).
El-Halfawy, O. M. & Valvano, M. A. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207 (2015). A comprehensive and clarifying review of heteroresistance in bacteria.
Alexander, H. E. & Leidy, G. Mode of action of streptomycin on type b Haemophilus influenzae: I. Origin of resistant organisms. J. Exp. Med. 85, 329–338 (1947).
Devi, Y., P. M., P., Thomas, S. & Veeraraghavan, B. Challenges in the laboratory diagnosis and clinical management of heteroresistant vancomycin Staphylococcus aureus (hVISA). Clin. Microbiol. 4, 214 (2015).
Zheng, C. et al. Mixed infections and rifampin heteroresistance among Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 53, 2138–2147 (2015).
Mascellino, M. T., Porowska, B., De Angelis, M. & Oliva, A. Antibiotic susceptibility, heteroresistance, and updated treatment strategies in Helicobacter pylori infection. Drug Des. Devel. Ther. 11, 2209–2220 (2017).
Canetti, G., Rist, N. & Grosset, J. Measurement of sensitivity of the tuberculous bacillus to antibacillary drugs by the method of proportions. Methodology, resistance criteria, results and interpretation [French]. Rev. Tuberc. Pneumol. 27, 217–272 (2015).
Barin, J., Martins, A. F., Heineck, B. L., Barth, A. L. & Zavascki, A. P. Hetero- and adaptive resistance to polymyxin B in OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolates. Ann. Clin. Microbiol. Antimicrob. 12, 15 (2013).
da Silva, A. E. B., Martins, A. F., Nodari, C. S., Magagnin, C. M. & Barth, A. L. Carbapenem-heteroresistance among isolates of the Enterobacter cloacae complex: is it a real concern? Eur. J. Clin. Microbiol. Infect. Dis. 37, 185–186 (2018).
Oikonomou, O., Panopoulou, M. & Ikonomidis, A. Investigation of carbapenem heteroresistance among different sequence types of Pseudomonas aeruginosa clinical isolates reveals further diversity. J. Med. Microbiol. 60, 1556–1558 (2011).
Hermes, D. M. et al. Evaluation of heteroresistance to polymyxin B among carbapenem-susceptible and -resistant Pseudomonas aeruginosa. J. Med. Microbiol. 62, 1184–1189 (2013).
Pournaras, S. et al. Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical Isolates of K. pneumoniae. J. Clin. Microbiol. 48, 2601–2604 (2010).
Nicoloff, H., Hjort, K., Levin, B. R. & Andersson, D. I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514 (2019). A comprehensive analysis of heteroresistance in four Gram-negative species that demonstrates that more than one quarter of all species–antibiotic combinations show heteroresistance and that it is mainly caused by gene amplification.
Hernan, R. C. et al. Selection of colistin-resistant Acinetobacter baumannii isolates in postneurosurgical meningitis in an intensive care unit with high presence of heteroresistance to colistin. Diagn. Microbiol. Infect. Dis. 65, 188–191 (2009).
Mei, S., Gao, Y., Zhu, C., Dong, C. & Chen, Y. Research of the heteroresistance of Pseudomonas aeruginosa to imipenem. Int. J. Clin. Exp. Med. 8, 6129–6132 (2015).
Wong, S. S. Y., Ho, P. L., Woo, P. C. Y. & Yuen, K. Y. Bacteremia caused by Staphylococci with inducible vancomycin heteroresistance. Clin. Infect. Dis. 29, 760–767 (1999).
Bardet, L. et al. Deciphering heteroresistance to colistin in a Klebsiella pneumoniae isolate from Marseille, France. Antimicrob. Agents Chemother. 61, e00356-17 (2017).
Lucas, A. E. et al. Frequency and mechanisms of spontaneous fosfomycin nonsusceptibility observed upon disk diffusion testing of Escherichia coli. J. Clin. Microbiol. 56, e01368-17 (2018).
Plipat, N., Livni, G., Bertram, H. & Thomson, R. B. Unstable vancomycin heteroresistance is common among clinical isolates of methiciliin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43, 2494–2496 (2005).
Anderson, S. E., Sherman, E. X., Weiss, D. S. & Rather, P. N. Aminoglycoside heteroresistance in Acinetobacter baumannii AB5075. mSphere 3, e00271-18 (2018). A study that demonstrates the importance of gene amplification in heteroresistance.
Trauner, A. et al. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 18, 71 (2017).
Lieberman, T. D. et al. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat. Med. 22, 1470–1474 (2016).
Drlica, K., Shopsin, B. & Zhao, X. in Antimicrobial Resistance in the 21st Century (eds Fong, I. W., Shlaes, D. & Drlica, K.) 269–296 (Springer, 2018).
Sun, L. et al. Droplet Digital PCR detection of Helicobacter pylori clarithromycin resistance reveals frequent heteroresistance. J. Clin. Microbiol. 56, e00019-18 (2018).
Colman, R. E. et al. Detection of low-level mixed-population drug resistance in Mycobacterium tuberculosis using high fidelity amplicon sequencing. PLOS ONE 10, e0126626 (2015).
Sanchez-Romero, M. A. & Casadesus, J. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc. Natl Acad. Sci. USA 111, 355–360 (2014).
Balaban, N. Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775 (2011).
Brauner, A., Fridman, O., Gefen, O. & Balaban, N. Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016).
Cannatelli, A. et al. MgrB inactivation is a common mechanism of colistin resistance in KPC carbapenemase-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703 (2014).
Machado, D. et al. Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J. Med. Microbiol. 67, 740–749 (2018).
He, J. et al. Heteroresistance to carbapenems in invasive Pseudomonas aeruginosa infections. Int. J. Antimicrob. Agents 51, 413–421 (2018).
Ikonomidis, A. et al. Efflux system overexpression and decreased OprD contribute to the carbapenem heterogeneity in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 279, 36–39 (2008).
Lee, H.-Y. et al. Imipenem heteroresistance induced by imipenem in multidrug-resistant Acinetobacter baumannii: mechanism and clinical implications. Int. J. Antimicrob. Agents 37, 302–308 (2011).
Fernández Cuenca, F. et al. Prevalence and analysis of microbiological factors associated with phenotypic heterogeneous resistance to carbapenems in Acinetobacter baumannii. Int. J. Antimicrob. Agents 39, 472–477 (2012).
Hjort, K., Nicoloff, H. & Andersson, D. I. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol. Microbiol. 102, 274–289 (2016). The first demonstration that heteroresistance can be caused by unstable gene amplifications.
Jayol, A., Nordmann, P., Brink, A. & Poirel, L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 59, 2780–2784 (2015).
Halaby, T. et al. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 60, 6837–6843 (2016).
Landman, D., Salamera, J. & Quale, J. Irreproducible and uninterpretable polymyxin B MICs for Enterobacter cloacae and Enterobacter aerogenes. J. Clin. Microbiol. 51, 4106–4111 (2013).
Engel, H. et al. Heteroresistance to fosfomycin is predominant in Streptococcus pneumoniae and depends on the murA1 gene. Antimicrob. Agents Chemother. 57, 2801–2808 (2013).
Higgins, P. G., Schneiders, T., Hamprecht, A. & Seifert, H. In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 gene into blaOXA-58 in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob. Agents Chemother. 54, 5021–5027 (2010).
Chen, Y. et al. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front. Cell. Infect. Microbiol. 7, 37 (2017).
Zheng, J. et al. Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg. Microbes Infect. 7, 139 (2018).
Thulin, E., Sundqvist, M. & Andersson, D. I. Amdinocillin (mecillinam) resistance mutations in clinical isolates and laboratory-selected mutants of Escherichia coli. Antimicrob. Agents Chemother. 59, 1718–1727 (2015).
Reams, A. B., Kofoid, E., Savageau, M. & Roth, J. R. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184, 1077–1094 (2010).
Anderson, P. & Roth, J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc. Natl Acad. Sci. USA 78, 3113–3117 (1981).
Pettersson, M. E., Sun, S., Andersson, D. I. & Berg, O. G. Evolution of new gene functions: simulation and analysis of the amplification model. Genetica 135, 309–324 (2009).
Reams, A. B. & Roth, J. R. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 7, a016592 (2015).
Sandegren, L. & Andersson, D. I. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 578–588 (2009).
Adler, M., Anjum, M., Berg, O. G., Andersson, D. I. & Sandegren, L. High fitness costs and instability of gene duplications reduce rates of evolution of new genes by duplication-divergence mechanisms. Mol. Biol. Evol. 31, 1526–1535 (2014).
Schechter, L. M. et al. Extensive gene amplification as a mechanism for piperacillin-tazobactam resistance in Escherichia coli. mBio 9, e00583-18 (2018).
Engel, H. et al. A low-affinity penicillin-binding protein 2x variant is required for heteroresistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 3934–3941 (2014).
Hooper, D. C. & Jacoby, G. A. Mechanisms of drug resistance: quinolone resistance. Ann. N. Y. Acad. Sci. 1354, 12–31 (2015).
Goldstein, B. P. Resistance to rifampicin: a review. J. Antibiot. 67, 625–630 (2014).
Lo-Ten-Foe, J. R., de Smet, A. M. G. A., Diederen, B. M. W., Kluytmans, J. A. J. W. & van Keulen, P. H. J. Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3726–3730 (2007).
van Hal, S. J. et al. Performance of various testing methodologies for detection of heteroresistant vancomycin-intermediate Staphylococcus aureus in bloodstream isolates. J. Clin. Microbiol. 49, 1489–1494 (2011).
Udekwu, K. I., Parrish, N., Ankomah, P., Baquero, F. & Levin, B. R. Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 63, 745–757 (2009).
Sautrey, G., Duval, R. E., Chevalley, A., Fontanay, S. & Clarot, I. Capillary electrophoresis for fast detection of heterogeneous population in colistin-resistant Gram-negative bacteria. Electrophoresis 36, 2630–2633 (2015).
Jayol, A. et al. Evaluation of the rapid polymyxin NP test and its industrial version for the detection of polymyxin-resistant Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 92, 90–94 (2018).
Asakura, K. et al. Rapid and easy detection of low-level resistance to vancomycin in methicillin-resistant Staphylococcus aureus by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLOS ONE 13, e0194212 (2018).
Price, C. S., Kon, S. E. & Metzger, S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J. Microbiol. Methods 98, 50–58 (2014).
Entenza, J. M. et al. Rapid detection of Staphylococcus aureus strains with reduced susceptibility to vancomycin by isothermal microcalorimetry. J. Clin. Microbiol. 52, 180–186 (2014).
Bradley, P. et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 6, 10063 (2015).
Pankhurst, L. J. et al. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir. Med. 4, 49–58 (2016).
Operario, D. J. et al. Prevalence and extent of heteroresistance by next generation sequencing of multidrug-resistant tuberculosis. PLOS ONE 12, e0176522 (2017).
Nunes, A. P. F. et al. Heterogeneous resistance to vancomycin and teicoplanin among Staphylococcus spp. isolated from bacteremia. Braz. J. Infect. Dis. 11, 345–350 (2007).
Juhász, E., Iván, M., Pintér, E., Pongrácz, J. & Kristóf, K. Colistin resistance among blood culture isolates at a tertiary care centre in Hungary. J. Glob. Antimicrob. Resist. 11, 167–170 (2017).
Zhang, S., Sun, X., Chang, W., Dai, Y. & Ma, X. Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLOS ONE 10, e0136082 (2015).
Park, Y. J. et al. Screening method for detecting staphylococci with reduced susceptibility to teicoplanin. J. Microbiol. Methods 40, 193–198 (2000).
Tevell, S., Claesson, C., Hellmark, B., Söderquist, B. & Nilsdotter-Augustinsson, Å. Heterogeneous glycopeptide intermediate Staphylococcus epidermidis isolated from prosthetic joint infections. Eur. J. Clin. Microbiol. Infect. Dis. 33, 911–917 (2014).
Chung, M. et al. Heterogeneous oxacillin-resistant phenotypes and production of PBP2A by oxacillin-susceptible/mecA-positive MRSA strains from Africa. J. Antimicrob. Chemother. 71, 2804–2809 (2016).
Frebourg, N. B., Nouet, D., Lemée, L., Martin, E. & Lemeland, J. F. Comparison of ATB staph, rapid ATB staph, Vitek, and E-test methods for detection of oxacillin heteroresistance in staphylococci possessing mecA. J. Clin. Microbiol. 36, 52–57 (1998).
Okado, J. B., Avaca-Crusca, J. S., Oliveira, A. L., Dabul, A. N. G. & Camargo, I. L. B. Daptomycin and vancomycin heteroresistance revealed among CC5-SCCmecII MRSA clone and in vitro evaluation of treatment alternatives. J. Glob. Antimicrob. Resist. 14, 209–216 (2018).
Sakoulas, G., Alder, J., Thauvin-Eliopoulos, C., Moellering, R. C. & Eliopoulos, G. M. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50, 1581–1585 (2006).
Saravolatz, S. N., Martin, H., Pawlak, J., Johnson, L. B. & Saravolatz, L. D. Ceftaroline-heteroresistant Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 3133–3136 (2014).
Coelho, C., de Lencastre, H. & Aires-de-Sousa, M. Frequent occurrence of trimethoprim-sulfamethoxazole hetero-resistant Staphylococcus aureus isolates in different African countries. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1243–1252 (2017).
Poudyal, A. et al. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 62, 1311–1318 (2008).
Meletis, G., Tzampaz, E., Sianou, E., Tzavaras, I. & Sofianou, D. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 66, 946–947 (2011).
Yau, W. et al. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J. Infect. 58, 138–144 (2009).
Guérin, F. et al. Cluster-dependent colistin hetero-resistance in Enterobacter cloacae complex. J. Antimicrob. Chemother. 71, 3058–3061 (2016).
Cai, Y., Chai, D., Wang, R., Liang, B. & Bai, N. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67, 1607–1615 (2012).
Walsh, C. C., McIntosh, M. P., Peleg, A. Y., Kirkpatrick, C. M. & Bergen, P. J. In vitro pharmacodynamics of fosfomycin against clinical isolates of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 70, 3042–3050 (2015).
Kaase, M., Szabados, F., Anders, A. & Gatermann, S. G. Fosfomycin susceptibility in carbapenem-resistant Enterobacteriaceae from Germany. J. Clin. Microbiol. 52, 1893–1897 (2014).
Ma, W., Sun, J., Yang, S. & Zhang, L. Epidemiological and clinical features for cefepime heteroresistant Escherichia coli infections in Southwest China. Eur. J. Clin. Microbiol. Infect. Dis. 35, 571–578 (2016).
Savini, V. et al. Misidentification of ampicillin-sulbactam heteroresistance in Acinetobacter baumannii strains from ICU patients. J. Infect. 58, 316–317 (2009).
Pelaez, T. et al. Metronidazole resistance in Clostridium difficile is heterogeneous. J. Clin. Microbiol. 46, 3028–3032 (2008).
Huang, H. et al. Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe 16, 633–635 (2010).
Álvarez-Pérez, S., Blanco, J. L., Harmanus, C., Kuijper, E. & García, M. E. Subtyping and antimicrobial susceptibility of Clostridium difficile PCR ribotype 078/126 isolates of human and animal origin. Vet. Microbiol. 199, 15–22 (2017).
Sun, J. D. et al. Impact of carbapenem heteroresistance among clinical isolates of invasive Escherichia coli in Chongqing, southwestern China. Clin. Microbiol. Infect. 21, 469.e1–469.e10 (2015).
Kang, C. K. et al. agr functionality affects clinical outcomes in patients with persistent methicillin-resistant Staphylococcus aureus bacteraemia. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2187–2191 (2017).
Park, K. H. et al. Comparison of the clinical features, bacterial genotypes and outcomes of patients with bacteraemia due to heteroresistant vancomycin-intermediate Staphylococcus aureus and vancomycin-susceptible S. aureus. J. Antimicrob. Chemother. 67, 1843–1849 (2012).
Maor, Y. et al. Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J. Infect. Dis. 199, 619–624 (2009).
Charles, P. G., Ward, P. B., Johnson, P. D., Howden, B. P. & Grayson, M. L. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38, 448–451 (2004).
Casapao, A. M. et al. Evaluation of vancomycin population susceptibility analysis profile as a predictor of outcomes for patients with infective endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 4636–4641 (2014).
Claeys, K. C. et al. Pneumonia caused by methicillin-resistant Staphylococcus aureus: Does vancomycin heteroresistance matter? Antimicrob. Agents Chemother. 60, 1708–1716 (2016).
van Hal, S. J., Jones, M., Gosbell, I. B. & Paterson, D. L. Vancomycin heteroresistance is associated with reduced mortality in ST239 methicillin-resistant Staphylococcus aureus blood stream infections. PLOS ONE 6, e21217 (2011).
Fernández-Cuenca, F. et al. Epidemiological and clinical features associated with colonisation/infection by Acinetobacter baumannii with phenotypic heterogeneous resistance to carbapenems. Int. J. Antimicrob. Agents 40, 235–238 (2012).
Di Gregorio, S. et al. Clinical, microbiological, and genetic characteristics of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia in a teaching hospital. Microb. Drug Resist. 21, 25–34 (2015).
Moosavian, M. et al. Post neurosurgical meningitis due to colistin heteroresistant Acinetobacter baumannii. Jundishapur J. Microbiol. 7, e12287 (2014).
Sieradzki, K., Roberts, R. B., Serur, D., Hargrave, J. & Tomasz, A. Heterogeneously vancomycin-resistant Staphylococcus epidermidis strain causing recurrent peritonitis in a dialysis patient during vancomycin therapy. J. Clin. Microbiol. 37, 39–44 (1999).
Rodríguez, C. H. et al. Impact of heteroresistance to colistin in meningitis caused by Acinetobacter baumannii. J. Infect. 64, 119–121 (2012).
Srinivas, P. et al. Detection of colistin heteroresistance in Acinetobacter baumannii from blood and respiratory isolates. Diagn. Microbiol. Infect. Dis. 91, 194–198 (2018).
Hiramatsu, K. et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350, 1670–1673 (1997).
Wootton, M. et al. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47, 399–403 (2001).
Band, V. I. et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat. Microbiol. 1, 16053 (2016). An important study demonstrating the ability of low-frequency bacterial subpopulations to contribute to clinically relevant colistin resistance.
Band, V. I. et al. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9, e02448-17 (2018). The first report of colistin-heteroresistant K. pneumoniae in the United States and a demonstration of how such isolates can lead to treatment failure in an in vivo model of infection.
Vaudaux, P., Francois, P., Berger-Bächi, B. & Lew, D. P. In vivo emergence of subpopulations expressing teicoplanin or vancomycin resistance phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphylococcus aureus. J. Antimicrob. Chemother. 47, 163–170 (2001).
Napier, B. A. et al. Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii. mBio 4, e00021-13 (2013).
Lippa, A. M. & Goulian, M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLOS Genet. 5, e1000788 (2009).
Cannatelli, A. et al. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526 (2013).
Moore, M. R., Perdreau-Remington, F. & Chambers, H. F. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47, 1262–1266 (2003).
Tan, C.-H., Li, J. & Nation, R. L. Activity of colistin against heteroresistant Acinetobacter baumannii and emergence of resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 51, 3413–3415 (2007).
Baltekin, Ö., Boucharin, A., Tano, E., Andersson, D. I. & Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl Acad. Sci. USA 114, 9170–9175 (2017).
Creely, D. et al. International dissemination of Escherichia coli strains with discrepant behaviour in phenotypic antimicrobial susceptibility tests. Eur. J. Clin. Microbiol. Infect. Dis. 32, 997–1002 (2013).
Shubert, C. et al. Population analysis of Escherichia coli isolates with discordant resistance levels by piperacillin-tazobactam broth microdilution and agar dilution testing. Antimicrob. Agents Chemother. 58, 1779–1781 (2014).
Ballestero-Téllez, M. et al. Role of inoculum and mutant frequency on fosfomycin MIC discrepancies by agar dilution and broth microdilution methods in Enterobacteriaceae. Clin. Microbiol. Infect. 23, 325–331 (2017).
Tato, M. et al. Carbapenem heteroresistance in VIM-1-producing Klebsiella pneumoniae isolates belonging to the same clone: consequences for routine susceptibility testing. J. Clin. Microbiol. 48, 4089–4093 (2010).
Hung, K.-H., Wang, M.-C., Huang, A.-H., Yan, J.-J. & Wu, J.-J. Heteroresistance to cephalosporins and penicillins in Acinetobacter baumannii. J. Clin. Microbiol. 50, 721–726 (2012).
Sun, S., Berg, O. G., Roth, J. R. & Andersson, D. I. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182, 1183–1195 (2009).
Li, J. et al. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 2946–2950 (2006).
Hawley, J. S., Murray, C. K. & Jorgensen, J. H. Colistin heteroresistance in acinetobacter and its association with previous colistin therapy. Antimicrob. Agents Chemother. 52, 351–352 (2008).
Cherkaoui, A. et al. Imipenem heteroresistance in nontypeable Haemophilus influenzae is linked to a combination of altered PBP3, slow drug influx and direct efflux regulation. Clin. Microbiol. Infect. 23, 118.e9–118.e19 (2017).
Pournaras, S. et al. Characterization of clinical isolates of Pseudomonas aeruginosa heterogeneously resistant to carbapenems. J. Med. Microbiol. 56, 66–70 (2007).
Pfeltz, R. F., Schmidt, J. L. & Wilkinson, B. J. A microdilution plating method for population analysis of antibiotic-resistant Staphylococci. Microb. Drug Resist. 7, 289–295 (2001).
Leonard, S. N., Rossi, K. L., Newton, K. L. & Rybak, M. J. Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J. Antimicrob. Chemother. 63, 489–492 (2009).
Satola, S. W., Farley, M. M., Anderson, K. F. & Patel, J. B. Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus, with the population analysis profile method as the reference method. J. Clin. Microbiol. 49, 177–183 (2011).
Silveira, A., Cunha, D., Caierão, G. R., Cordova, D. & Azevedo, C. Evaluation of the accuracy of phenotypic methods for the detection of heteroresistant vancomycin-intermediate Staphylococcus aureus (hVISA). JSM Microbiol. 4, 1031 (2016).
Gordon, N. C. & Wareham, D. W. Failure of the MicroScan WalkAway system to detect heteroresistance to carbapenems in a patient with Enterobacter aerogenes bacteremia. J. Clin. Microbiol. 47, 3024–3025 (2009).
Zhang, Z., Wang, Y., Pang, Y. & Liu, C. Comparison of different drug susceptibility test methods to detect rifampin heteroresistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 5632–5635 (2014).
Zhang, Z., Lu, J., Wang, Y., Pang, Y. & Zhao, Y. Automated liquid culture system misses isoniazid heteroresistance in Mycobacterium tuberculosis isolates with mutations in the promoter region of the inhA gene. Eur. J. Clin. Microbiol. Infect. Dis. 34, 555–560 (2015).
Nikolayevskyy, V. et al. Performance of the Genotype® MTBDRPlus assay in the diagnosis of tuberculosis and drug resistance in Samara, Russian Federation. BMC Clin. Pathol. 9, 2 (2009).
Rinder, H., Mieskes, K. T. & Löscher, T. Heteroresistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 5, 339–345 (2001).
Chakravorty, S. et al. The Nnew Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. mBio 8, e00812-17 (2017).
Driesen, M. et al. Evaluation of a novel line probe assay to detect resistance to pyrazinamide, a key drug used for tuberculosis treatment. Clin. Microbiol. Infect. 24, 60–64 (2018).
Hofmann-Thiel, S. et al. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur. Respir. J. 33, 368–374 (2008).
Cambau, E. et al. Evaluation of a new test, GenoType HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J. Clin. Microbiol. 47, 3600–3607 (2009).
Brennan, D. E. et al. Molecular detection of Helicobacter pylori antibiotic resistance in stool versus biopsy samples. World J. Gastroenterol. 22, 9214–9221 (2016).
Zetola, N. M. et al. Mixed Mycobacterium tuberculosis complex infections and false-negative results for rifampin resistance by GeneXpert MTB/RIF are associated with poor clinical outcomes. J. Clin. Microbiol. 52, 2422–2429 (2014).
Acknowledgements
Work in the authors’ laboratory was supported by a grant from the Swedish Research Council (to D.I.A.).
Author information
Authors and Affiliations
Contributions
D.I.A., H.N. and K.H. researched data for the article, contributed substantially to the discussion of content, wrote the article and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information
Nature Reviews Microbiology thanks W. van Schaik, M. Valvano and D. S. Weiss for their contribution to the peer review of this work.
Glossary
- Minimal inhibitory concentration
-
(MIC). The lowest concentration of an antimicrobial drug that prevents growth of a bacterial population.
- Antimicrobial susceptibility test
-
(AST). A method used to determine the minimal inhibitory concentration or clinical resistance level of a bacterial isolate.
- Clinical breakpoint
-
The concentration of an antibiotic that defines whether infection with a bacterial species is treatable with the antibiotic (susceptible) or untreatable (resistant) with the antibiotic.
- Persister cells
-
Bacteria that are sensitive to an antibiotic but that can survive in the presence of the antibiotic because their growth has temporarily stopped.
- Compensatory mutations
-
Mutations that alleviate or suppress the phenotypic effect of a previous mutation.
- Small colony variant
-
Naturally occurring bacterial mutants with defects in electron transport that result in slow growth and resistance to certain antibiotic classes.
- Pseudorevertants
-
Mutants that carry a second mutation (see Compensatory mutations) that alleviates or reverses the phenotypic effects of an already existing mutation.
- Inoculum effect
-
A phenomenon where the minimal inhibitory concentration of an antibiotic increases with the density of cells in the inoculum in the antimicrobial susceptibility test.
- Major errors
-
Major errors are observed when one bacterial isolate is characterized as susceptible to an antibiotic when tested by one type of antimicrobial susceptibility test but is characterized as resistant to that antibiotic when tested by another antimicrobial susceptibility test.
Rights and permissions
About this article
Cite this article
Andersson, D.I., Nicoloff, H. & Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol 17, 479–496 (2019). https://doi.org/10.1038/s41579-019-0218-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-019-0218-1
This article is cited by
-
Bacteria can compensate the fitness costs of amplified resistance genes via a bypass mechanism
Nature Communications (2024)
-
Drug-resistant tuberculosis: a persistent global health concern
Nature Reviews Microbiology (2024)
-
Plasmid-mediated phenotypic noise leads to transient antibiotic resistance in bacteria
Nature Communications (2024)
-
Emerging tools for uncovering genetic and transcriptomic heterogeneities in bacteria
Biophysical Reviews (2024)
-
Multiple heteroresistance to tigecycline and colistin in Acinetobacter baumannii isolates and its implications for combined antibiotic treatment
Journal of Biomedical Science (2023)