Abstract
Phages differ substantially in the bacterial hosts that they infect. Their host range is determined by the specific structures that they use to target bacterial cells. Tailed phages use a broad range of receptor-binding proteins, such as tail fibres, tail spikes and the central tail spike, to target their cognate bacterial cell surface receptors. Recent technical advances and new structure–function insights have begun to unravel the molecular mechanisms and temporal dynamics that govern these interactions. Here, we review the current understanding of the targeting machinery and mechanisms of tailed phages. These new insights and approaches pave the way for the application of phages in medicine and biotechnology and enable deeper understanding of their ecology and evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


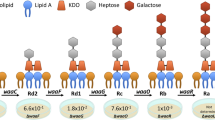
Adapted with permission from ref.4, Proceedings of the National Academy of Sciences of the United States of America.


Similar content being viewed by others
References
Burns, C. M., Chan, H. L. & DuBow, M. S. In vitro maturation and encapsidation of the DNA of transposable Mu-like phage D108. Proc. Natl Acad. Sci. USA 87, 6092–6096 (1990).
Stockley, P. G. et al. Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid. Bacteriophage 6, e1157666 (2016).
Ackermann, H.-W. & Prangishvili, D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 157, 1843–1849 (2012).
Hu, B., Margolin, W., Molineux, I. J. & Liu, J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc. Natl Acad. Sci. USA 112, E4919–E4928 (2015). This study analyses cryo-electron microscopic images of phage T4 at different stages of infection to reveal that, contrary to common descriptions, most long tail fibres are folded back against the virion before infection.
Arnaud, C.-A. et al. Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection. Nat. Commun. 8, 1953 (2017).
González-García, V. A. et al. Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J. Biol. Chem. 290, 10038–10044 (2015).
Taylor, N. M. I. et al. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533, 346–352 (2016). This study presents the atomic structure of the phage T4 baseplate before and after attachment states, detailing the events leading to sheath contraction at the atomic level.
Keen, E. C. & Adhya, S. L. Review of phage therapy: current research and applications. Clin. Infect. Dis. 61, 141–142 (2015).
Motlagh, A. M. et al. Insights of phage-host interaction in hypersaline ecosystem through metagenomics analyses. Front. Microbiol. 8, 352 (2017).
Leiman, P. G. & Shneider, M. Contractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 726, 93–114 (2012).
Leiman, P. G. et al. Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355–355 (2010).
Kellenberger, E. et al. Functions and properties related to the tail fibers of bacteriophage T4. Virology 26, 419–440 (1965).
Kostyuchenko, V. A. et al. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 12, 810 (2005).
Tétart, F., Repoila, F., Monod, C. & Krisch, H. M. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J. Mol. Biol. 258, 726–731 (1996).
Ge, P. et al. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat. Struct. Mol. Biol. 22, 377–382 (2015).
Büttner, C. R., Wu, Y., Maxwell, K. L. & Davidson, A. R. Baseplate assembly of phage Mu: defining the conserved core components of contractile-tailed phages and related bacterial systems. Proc. Natl Acad. Sci. USA 113, 10174–10179 (2016). This study defines the assembly pathway of the phage Mu baseplate and demonstrates that Mu-like, simple baseplates are conserved in prophages and are the most likely evolutionary precursors of contractile, tail-derived bacterial apparatuses, such as the T6SS.
Nováček, J. et al. Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc. Natl Acad. Sci. USA 113, 9351–9356 (2016).
Chibani-Chennoufi, S., Dillmann, M.-L., Marvin-Guy, L., Rami-Shojaei, S. & Brüssow, H. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 186, 7069–7083 (2004).
Kutter, E. M. et al. Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol. J. 8, 430–430 (2011).
Arachchi, G. J. G. et al. Characteristics of three listeriaphages isolated from New Zealand seafood environments. J. Appl. Microbiol. 115, 1427–1438 (2013).
Fokine, A. & Rossmann, M. G. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 4, e28281 (2014).
Zivanovic, Y. et al. Insights into bacteriophage T5 structure from analysis of its morphogenesis genes and protein components. J. Virol. 88, 1162–1174 (2014).
Böhm, J. et al. FhuA-mediated phage genome transfer into liposomes. Curr. Biol. 11, 1168–1175 (2001).
Flayhan, A. et al. Crystal structure of pb9, the distal tail protein of bacteriophage T5: a conserved structural motif among all siphophages. J. Virol. 88, 820–828 (2014).
Golomidova, A. et al. Branched lateral tail fiber organization in T5-like bacteriophages DT57C and DT571/2 is revealed by genetic and functional analysis. Viruses 8, 26 (2016).
Scholl, D., Rogers, S., Adhya, S. & Merril, C. R. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75, 2509–2515 (2001).
Prokhorov, N. S. et al. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 105, 385–398 (2017).
Farenc, C. et al. Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J. Virol. 88, 7005–7015 (2014).
Sciara, G. et al. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl Acad. Sci. USA 107, 6852–6857 (2010).
Bebeacua, C. et al. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J. Virol. 87, 12302–12312 (2013).
McCabe, O. et al. The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol. Microbiol. 96, 875–886 (2015).
Mahony, J., Randazzo, W., Neve, H., Settanni, L. & van Sinderen, D. Lactococcal 949 group phages recognize a carbohydrate receptor on the host cell surface. Appl. Environ. Microbiol. 81, 3299–3305 (2015).
Cuervo, A. et al. Structural characterization of the bacteriophage T7 tail machinery. J. Biol. Chem. 288, 26290–26299 (2013).
Steinbacher, S. et al. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267, 865–880 (1997).
Israel, V. A model for the adsorption of phage P22 to Salmonella typhimurium. J. Gen. Virol. 40, 669–673 (1978).
Olia, A. S., Casjens, S. & Cingolani, G. Structure of phage P22 cell envelope-penetrating needle. Nat. Struct. Mol. Biol. 14, 1221–1226 (2007).
Olia, A. S., Casjens, S. & Cingolani, G. Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas. Protein Sci. 18, 537–548 (2009).
Perez, G. L., Huynh, B., Slater, M. & Maloy, S. Transport of phage P22 DNA across the cytoplasmic membrane. J. Bacteriol. 191, 135–140 (2009).
Tang, J. et al. DNA poised for release in bacteriophage ø29. Structure 16, 935–943 (2008). This study presents the first asymmetrical 3D reconstruction of a tailed double-stranded DNA phage at subnanometre resolution, providing important insights into the interactions of protein and DNA components during phage assembly.
Xiang, Y. et al. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 34, 375–386 (2009).
Xu, J., Gui, M., Wang, D. & Xiang, Y. The bacteriophage φ29 tail possesses a pore-forming loop for cell membrane penetration. Nature 534, 544–547 (2016). This study demonstrates that prokaryotic and eukaryotic viruses use similar mechanisms to breach the cell membrane. As in some non-enveloped eukaryotic viruses, phage φ29 forms a pore with six hydrophobic loops that form a channel through the membrane and unblock the tail tube to enable DNA ejection.
Chibani-Chennoufi, S., Bruttin, A., Dillmann, M.-L. & Brüssow, H. Phage-host interaction: an ecological perspective. J. Bacteriol. 186, 3677–3686 (2004).
Taylor, N. M. I., van Raaij, M. J. & Leiman, P. G. Contractile injection systems of bacteriophages and related systems. Mol. Microbiol. 108, 6–15 (2018).
Andres, D. et al. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J. Biol. Chem. 285, 36768–36775 (2010).
Müller, J. J. et al. An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri phage Sf6. Structure 16, 766–775 (2008).
Tang, L., Marion, W. R., Cingolani, G., Prevelige, P. E. & Johnson, J. E. Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J. 24, 2087–2095 (2005).
Seul, A. et al. Bacteriophage P22 tailspike: structure of the complete protein and function of the interdomain linker. Acta Cryst. D 70, 1336–1345 (2014). This study resolves the crystal structure of the phage P22 tail spike by making a single amino-acid exchange in the linker sequence between the particle binding domain and the receptor binding domain of the protein. It also suggests a role of the linker in signal transmission from the distal tip of the tail spike to the phage head, eventually leading to DNA ejection.
Barbirz, S. et al. Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol. Microbiol. 69, 303–316 (2008).
Dobbins, A. T. et al. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J. Bacteriol. 186, 1933–1944 (2004).
Walter, M. et al. Structure of the receptor-binding protein of bacteriophage Det7: a podoviral tail spike in a myovirus. J. Virol. 82, 2265–2273 (2008).
Olszak, T. et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci. Rep. 7, 16302 (2017).
Pickard, D. et al. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J. Bacteriol. 192, 5746–5754 (2010).
Szczeklik, H. & Taylor, A. The presence of Vi-polysaccharide deacetylase in three morphologically different Vi-bacteriophages. Biul. Inst. Med. Morsk. Gdansk. 24, 165–167 (1973).
King, J. & Laemmli, U. Polypeptides of the tail fibres of bacteriophage T4. J. Mol. Biol. 62, 465–477 (1971).
Cerritelli, M. E., Wall, J. S., Simon, M. N., Conway, J. F. & Steven, A. C. Stoichiometry and domainal organization of the long tail-fiber of bacteriophage T4: a hinged viral adhesin. J. Mol. Biol. 260, 767–780 (1996).
Hashemolhosseini, S., Stierhof, Y. D., Hindennach, I. & Henning, U. Characterization of the helper proteins for the assembly of tail fibers of coliphages T4 and lambda. J. Bacteriol. 178, 6258–6265 (1996).
Bartual, S. G. et al. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc. Natl Acad. Sci. USA 107, 20287–20292 (2010).
Washizaki, A., Yonesaki, T. & Otsuka, Y. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. MicrobiologyOpen 5, 1003–1015 (2016).
Hendrix, R. & Duda, R. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science 258, 1145–1148 (1992).
van Raaij, M. J., Schoehn, G., Burda, M. R. & Miller, S. Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J. Mol. Biol. 314, 1137–1146 (2001).
Mason, W. S. & Haselkorn, R. Product of T4 gene 12. J. Mol. Biol. 66, 445–469 (1972).
Thomassen, E. et al. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J. Mol. Biol. 331, 361–373 (2003).
Burda, M. R. & Miller, S. Folding of coliphage T4 short tail fiber in vitro. Eur. J. Biochem. 265, 771–778 (1999).
Leiman, P. G., Shneider, M. M., Mesyanzhinov, V. V. & Rossmann, M. G. Evolution of bacteriophage tails: structure of T4 gene product 10. J. Mol. Biol. 358, 912–921 (2006).
Garcia-Doval, C., Luque, D., Castón, J. R., Boulanger, P. & van Raaij, M. J. Crystallization of the C-terminal domain of the bacteriophage T5 L-shaped fibre. Acta Cryst. F 69, 1363–1367 (2013).
Schulz, E. C. & Ficner, R. Knitting and snipping: chaperones in β-helix folding. Curr. Opin. Struct. Biol. 21, 232–239 (2011).
Garcia-Doval, C. et al. Structure of the receptor-binding carboxy-terminal domain of the bacteriophage T5 L-shaped tail fibre with and without its intra-molecular chaperone. Viruses 7, 6424–6440 (2015).
Spinelli, S., Veesler, D., Bebeacua, C. & Cambillau, C. Structures and host-adhesion mechanisms of lactococcal siphophages. Front. Microbiol. 5, 3 (2014).
Garcia-Doval, C. & van Raaij, M. J. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc. Natl Acad. Sci. USA 109, 9390–9395 (2012).
Veesler, D. & Cambillau, C. A. Common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75, 423–433 (2011).
Pires, D. P., Oliveira, H., Melo, L. D. R., Sillankorva, S. & Azeredo, J. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100, 2141–2151 (2016).
Kanamaru, S. et al. Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553 (2002).
Nakagawa, H., Arisaka, F. & Ishii, S. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J. Virol. 54, 460–466 (1985).
Boulanger, P. et al. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J. Biol. Chem. 283, 13556–13564 (2008).
Cumby, N., Reimer, K., Mngin-Lecreulx, D., Davidson, A. & Maxwell, K. The phage tail tape measure protein, an inner membrane protein and a periplasmid chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol. Microbiol. 96, 437–447 (2015).
Bhardwaj, A., Olia, A. S., Walker-Kopp, N. & Cingolani, G. Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. J. Mol. Biol. 371, 374–387 (2007).
Xiang, Y. et al. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage ϕ29 tail. Proc. Natl Acad. Sci. USA 105, 9552–9557 (2008).
Ray, A. et al. Type VI secretion system MIX-effectors carry both antibacterial and anti-eukaryotic activities. EMBO Rep. 18, 1978–1990 (2017).
Yang, G., Dowling, A. J., Gerike, U., ffrench-Constant, R. H. & Waterfield, N. R. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J. Bacteriol. 188, 2254–2261 (2006).
Berkane, E. et al. Interaction of bacteriophage lambda with its cell surface receptor: an in vitro study of binding of the viral tail protein gpJ to LamB (maltoporin). Biochemistry 45, 2708–2720 (2006).
Stockdale, S. R. et al. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J. Biol. Chem. 288, 5581–5590 (2013).
Stamereilers, C., LeBlanc, L., Yost, D., Amy, P. S. & Tsourkas, P. K. Comparative genomics of 9 novel Paenibacillus larvae bacteriophages. Bacteriophage 6, e1220349 (2016).
Ceyssens, P. et al. Comparative analysis of the widespread and conserved PB1-like viruses infecting Pseudomonas aeruginosa. Environ. Microbiol. 11, 2874–2883 (2009).
Garbe, J. et al. Characterization of JG024, a Pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 10, 1–10 (2010).
Li, L. et al. Neutron crystallographic studies of T4 lysozyme at cryogenic temperature. Protein Sci. 26, 2098–2104 (2017).
Oliveira, H. et al. Characterization and genomic analyses of two newly isolated Morganella phages define distant members among Tevenvirinae and Autographivirinae subfamilies. Sci. Rep. 7, 46157 (2017).
Koraimann, G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60, 2371–2388 (2003).
Piuri, M. & Hatfull, G. F. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62, 1569–1585 (2006).
Kanamaru, S., Ishiwata, Y., Suzuki, T., Rossmann, M. G. & Arisaka, F. Control of bacteriophage T4 tail lysozyme activity during the infection process. J. Mol. Biol. 346, 1013–1020 (2005).
Glonti, T., Chanishvili, N. & Taylor, P. W. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol. 108, 695–702 (2010).
Majkowska-Skrobek, G. et al. Capsule-targeting depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses 8, 324 (2016).
Hamdi, S. et al. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci. Rep. 7, 40349 (2017).
Gutiérrez, D. et al. Role of the pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in staphylococcal species. Front. Microbiol. 6, 1315 (2015).
Rakhuba, D., Kolomiets, E., Dey, E. & Novik, G. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59, 145–155 (2010).
Köhler, T., Donner, V. & van Delden, C. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J. Bacteriol. 192, 1921–1928 (2010).
Kim, M., Kim, S., Park, B. & Ryu, S. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella Biocontrol. Appl. Environ. Microbiol. 80, 1026–1034 (2014).
Zhao, X. et al. Outer membrane proteins ail and OmpF of Yersinia pestis are involved in the adsorption of T7-related bacteriophage Yep-phi. J. Virol. 87, 12260–12269 (2013).
Kim, M. & Ryu, S. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 86, 411–425 (2012).
Gehring, K., Charbit, A., Brissaud, E. & Hofnung, M. Bacteriophage lambda receptor site on the Escherichia coli K-12 LamB protein. J. Bacteriol. 169, 2103–2106 (1987).
Ricci, V. & Piddock, L. J. V. Exploiting the role of TolC in pathogenicity: identification of a bacteriophage for eradication of Salmonella serovars from poultry. Appl. Environ. Microbiol. 76, 1704–1706 (2010).
Breyton, C. et al. Assessing the conformational changes of pb5, the receptor-binding protein of phage T5, upon binding to its Escherichia coli receptor FhuA. J. Biol. Chem. 288, 30763–30772 (2013).
Rabsch, W. et al. FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J. Bacteriol. 189, 5658–5674 (2007).
Rakieten, M. & Rakieten, T. Relationship between staphylococci and bacilli belonging to the subtilis group as shown by bacteriophage adsorption. J. Bacteriol. 34, 285–300 (1937).
Baptista, C., Santos, M. A. & São-José, C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J. Bacteriol. 190, 4989–4996 (2008).
Xia, G. et al. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 193, 4006–4009 (2011).
Li, X. et al. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci. Rep. 6, 26455 (2016).
Davison, S., Couture-Tosi, E., Candela, T., Mock, M. & Fouet, A. Identification of the Bacillus anthracis γ phage receptor. J. Bacteriol. 187, 6742–6749 (2005).
São-José, C. et al. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 281, 11464–11470 (2006).
Monteville, M. R., Ardestani, B. & Geller, B. L. Lactococcal bacteriophages require a host cell wall carbohydrate and a plasma membrane protein for adsorption and ejection of DNA. Appl. Environ. Microbiol. 60, 3204–3211 (1994).
Tzipilevich, E., Habusha, M. & Ben-Yehuda, S. Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168, 186–199 (2017).
Guerrero-Ferreira, R. C. et al. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl Acad. Sci. USA 108, 9963–9968 (2011). This study demonstrates that some phages use a filament on the phage head to interact with the bacterial flagellum and thus concentrate phage particles around the receptor on the cell surface; the study suggests that similar mechanisms apply to other phages and hosts.
Choi, Y., Shin, H., Lee, J.-H. & Ryu, S. Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl. Environ. Microbiol. 79, 4829–4837 (2013).
Raimondo, L. M., Lundh, N. P. & Martinez, R. J. Primary adsorption site of phage PBS1: the flagellum of Bacillus subtilis. J. Virol. 2, 256–264 (1968).
Lovett, P. S. PBP1: a flagella specific bacteriophage mediating transduction in Bacillus pumilus. Virology 47, 743–752 (1972).
Evans, T. J. et al. Characterization of a broad-host-range flagellum-dependent phage that mediates high-efficiency generalized transduction in, and between, Serratia and Pantoea. Microbiology 156, 240–247 (2010).
Evans, T. J., Trauner, A., Komitopoulou, E. & Salmond, G. P. C. Exploitation of a new flagellatropic phage of Erwinia for positive selection of bacterial mutants attenuated in plant virulence: towards phage therapy. J. Appl. Microbiol. 108, 676–685 (2009).
Pate, J. L., Petzold, S. J. & Umbreit, T. H. Two flagellotropic phages and one pilus-specific phage active against Asticcacaulis biprosthecum. Virology 94, 24–37 (1979).
Zhang, H., Li, L., Zhao, Z., Peng, D. & Zhou, X. Polar flagella rotation in Vibrio parahaemolyticus confers resistance to bacteriophage infection. Sci. Rep. 6, 26147 (2016).
Chibeu, A. et al. The adsorption of Pseudomonas aeruginosa bacteriophage ϕKMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol. Lett. 296, 210–218 (2009).
Bae, H.-W. & Cho, Y.-H. Complete genome sequence of Pseudomonas aeruginosa podophage MPK7, which requires type IV pili for infection. Genome Announc. 1, e00744–00713 (2013).
Costa & Tiago, R. et al. Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell 166, 1436–1444 (2016).
Frost, L. & Paranchych, W. DNA sequence analysis of point mutations in traA, the F pilin gene, reveal two domains involved in F-specific bacteriophage attachment. Mol. Gen. Genet. 213, 134–139 (1988).
Harvey, H. et al. Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation. Nat. Microbiol. 3, 47–52 (2018).
Hsu, C.-R., Lin, T.-L., Pan, Y.-J., Hsieh, P.-F. & Wang, J.-T. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLOS ONE 8, e70092 (2013).
Scholl, D., Adhya, S. & Merril, C. Escherichia coli K1’s capsule is a barrier to bacteriophage T7. Appl. Environ. Microbiol. 71, 4872–4874 (2005).
Oliveira, H. et al. Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ. Microbiol. 19, 5060–5077 (2017).
Ozaki, T., Abe, N., Kimura, K., Suzuki, A. & Kaneko, J. Genomic analysis of Bacillus subtilis lytic bacteriophage φNIT1 capable of obstructing natto fermentation carrying genes for the capsule-lytic soluble enzymes poly-γ-glutamate hydrolase and levanase. Biosci. Biotechnol. Biochem. 81, 135–146 (2017).
Lee, K. K. & Gui, L. Dissecting virus infectious cycles by cryo-electron microscopy. PLOS Pathog. 12, e1005625 (2016). This study elucidates the mechanism of genome ejection of a Twort-like myophage: tail sheath contraction induces conformational changes to release the DNA from the connector complex; the DNA enters the neck; and further structural change in the tail tube enables genome release.
Orlov, I. & Klaholz, B. in Europ. Microsc. Congress 2016 Proc. 27–28 (2016).
Doerr, A. Single-particle cryo-electron microscopy. Nat. Methods 13, 23–23 (2016).
Kauffman, K. M. et al. A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554, 118 (2018).
Schooley, R. T. et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954–e00917 (2017).
Zhvania, P., Hoyle, N. S., Nadareishvili, L., Nizharadze, D. & Kutateladze, M. Phage therapy in a 16-year-old boy with Netherton syndrome. Front. Med. 4, 94 (2017).
Abedon, S. in Phage Therapy: Current Research and Applications Ch. 3 (eds Borysowski, J., Międzybrodzki, R. & Górski, A.) (Caister Academic Press, 2014).
Cui, L. & Bikard, D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 44, 4243–4251 (2016).
Nobrega, F. L. et al. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci. Rep. 6, 39235 (2016).
Das, M., Bhowmick, T. S., Ahern, S. J., Young, R. & Gonzalez, C. F. Control of Pierce’s disease by phage. PLOS ONE 10, e0128902 (2015).
Wang, Y. et al. Bacteriophage therapy for the control of Vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 473, 251–258 (2017).
Bao, H. et al. Bio-control of Salmonella enteritidis in foods using bacteriophages. Viruses 7, 4836–4853 (2015).
Mortari, A., Adami, A. & Lorenzelli, L. An unconventional approach to impedance microbiology: detection of culture media conductivity variations due to bacteriophage generated lyses of host bacteria. Biosens. Bioelectron. 67, 615–620 (2015).
Chen, J., Alcaine, S. D., Jiang, Z., Rotello, V. M. & Nugen, S. R. Detection of Escherichia coli in drinking water using T7 bacteriophage-conjugated magnetic probe. Anal. Chem. 87, 8977–8984 (2015).
Sergueev, K. V., Filippov, A. A. & Nikolich, M. P. Highly sensitive bacteriophage-based detection of Brucella abortus in mixed culture and spiked blood. Viruses 9, 144 (2017).
McNerney, R., Kambashi, B. S., Kinkese, J., Tembwe, R. & Godfrey-Faussett, P. Development of a bacteriophage phage replication assay for diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 42, 2115–2120 (2004).
Vinay, M. et al. Phage-based fluorescent biosensor prototypes to specifically detect enteric bacteria such as E. coli and Salmonella enterica Typhimurium. PLOS ONE 10, e0131466 (2015).
Arya, S. K. et al. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst 136, 486–492 (2011).
Srivastava, S. K. et al. Highly sensitive and specific detection of E. coli by a SERS nanobiosensor chip utilizing metallic nanosculptured thin films. Analyst 140, 3201–3209 (2015).
Moghtader, F., Congur, G., Zareie, H. M., Erdem, A. & Piskin, E. Impedimetric detection of pathogenic bacteria with bacteriophages using gold nanorod deposited graphite electrodes. RSC Adv. 6, 97832–97839 (2016).
Janczuk, M. et al. Bacteriophage-based bioconjugates as a flow cytometry probe for fast bacteria detection. Bioconjug. Chem. 28, 419–425 (2017).
Wang, Z., Wang, D., Kinchla, A. J., Sela, D. A. & Nugen, S. R. Rapid screening of waterborne pathogens using phage-mediated separation coupled with real-time PCR detection. Anal. Bioanal. Chem. 408, 4169–4178 (2016).
Zhang, Y., Yan, C., Yang, H., Yu, J. & Wei, H. Rapid and selective detection of E. coli O157:H7 combining phagomagnetic separation with enzymatic colorimetry. Food Chem. 234, 332–338 (2017).
Tay, L.-L. et al. Silica encapsulated SERS nanoprobe conjugated to the bacteriophage tailspike protein for targeted detection of Salmonella. Chem. Comm. 48, 1024–1026 (2012).
Schmidt, A., Rabsch, W., Broeker, N. K. & Barbirz, S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 16, 207 (2016).
Pan, Y.-J. et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob. Agents Chemother. 59, 1038–1047 (2015).
Lin, T.-L. et al. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 210, 1734–1744 (2014).
Hsieh, P.-F., Lin, H.-H., Lin, T.-L., Chen, Y.-Y. & Wang, J.-T. Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: Isolation and functional characterization. Sci. Rep. 7, 4624 (2017).
Nakayama, K. et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38, 213–231 (2000).
Scholl, D. Phage tail-like bacteriocins. Annu. Rev. Virol. 4, 453–467 (2017). This review provides a comprehensive overview of the function, structure and engineering of antibacterial tailocins that are released by cell lysis.
Uratani, Y. & Hoshino, T. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157, 632–636 (1984).
Sarris, P. F., Ladoukakis, E. D., Panopoulos, N. J. & Scoulica, E. V. A phage tail-derived element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol. Evol. 6, 1739–1747 (2014).
Hurst, M. R., Beard, S. S., Jackson, T. A. & Jones, S. M. Isolation and characterization of the Serratia entomophila antifeeding prophage. FEMS Microbiol. Lett. 270, 42–48 (2007).
Shikuma, N. J. et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343, 529–533 (2014).
Leiman, P. G. et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl Acad. Sci. USA 106, 4154–4159 (2009).
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y. & Attree, I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10, 104 (2009).
Chang, Y. W., Rettberg, L. A., Ortega, D. R. & Jensen, G. J. In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 18, 1090–1099 (2017).
Gallique, M., Bouteiller, M. & Merieau, A. The type VI secretion system: a dynamic system for bacterial communication? Front. Microbiol. 8, 1454 (2017).
Kohler, T., Donner, V. & van Delden, C. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J. Bacteriol. 192, 1921–1928 (2010).
Williams, S. R., Gebhart, D., Martin, D. W. & Scholl, D. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74, 3868–3876 (2008). This innovative paper describes how the host range of R-type pyocins can be reprogrammed by replacing parts of the tail fibres between phages with different host ranges.
Gebhart, D. et al. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio 6, e02368 (2015).
Steinbacher, S. et al. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265, 383–386 (1994).
Heller, K. & Braun, V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 139, 32–38 (1979).
Wang, J., Hofnung, M. & Charbit, A. The c-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182, 508–512 (2000).
German, G. J. & Misra, R. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage1. J. Mol. Biol. 308, 579–585 (2001).
Schade, S. Z., Adler, J. & Ris, H. How bacteriophage χ attacks motile bacteria. J. Virol. 1, 599–609 (1967).
Budzik, J. M., Rosche, W. A., Rietsch, A. & O’Toole, G. A. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 186, 3270–3273 (2004).
Munsch-Alatossava, P. & Alatossava, T. The extracellular phage-host interactions involved in the bacteriophage LL-H infection of Lactobacillus delbrueckii ssp. lactis ATCC 15808. Front. Microbiol. 4, 408 (2013).
Andres, D. et al. Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system. Mol. Microbiol. 83, 1244–1253 (2012).
Vinga, I. et al. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 83, 289–303 (2012).
Bradbeer, C., Woodrow, M. L. & Khalifah, L. I. Transport of vitamin B12 in Escherichia coli: common receptor system for vitamin B12 and bacteriophage BF23 on the outer membrane of the cell envelope. J. Bacteriol. 125, 1032–1039 (1976).
Heller, K. J. & Schwarz, H. Irreversible binding to the receptor of bacteriophages T5 and BF23 does not occur with the tip of the tail. J. Bacteriol. 162, 621–625 (1985).
Sandulache, R., Prehm, P., Expert, D., Toussaint, A. & Kamp, D. The cell wall receptor for bacteriophage Mu G(−) in Erwinia and Escherichia coli C. FEMS Microbiol. Lett. 28, 307–310 (1985).
Yoichi, M., Abe, M., Miyanaga, K., Unno, H. & Tanji, Y. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J. Biotechnol. 115, 101–107 (2005).
Lee, J.-H., Shin, H., Kim, H. & Ryu, S. Complete genome sequence of Salmonella bacteriophage SPN3US. J. Virol. 85, 13470–13471 (2011).
Habann, M. et al. Listeria phage A511, a model for the contractile tail machineries of SPO1-related bacteriophages. Mol. Microbiol. 92, 84–99 (2014).
Matilla, M. A. & Salmond, G. P. C. Bacteriophage φMAM1, a Viunalikevirus, is a broad-host-range, high-efficiency generalized transducer that infects environmental and clinical isolates of the enterobacterial genera Serratia and Kluyvera. Appl. Environ. Microbiol. 80, 6446–6457 (2014).
Prehm, P., Jann, B., Jann, K., Schmidt, G. & Stirm, S. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coli B. J. Mol. Biol. 101, 277–281 (1976).
Parent, K. N. et al. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol. Microbiol. 92, 47–60 (2014).
Acknowledgements
F.L.N. is supported by the Netherlands Organization for Scientific Research (NWO) Veni grant 016.Veni.181.092. P.A.d.J. and B.E.D. are supported by NWO Vidi grant 864.14.004. H.J.E.B. and L.L.D. are supported by the NWO/OCW as part of the Frontiers of Nanoscience programme. R.L. is supported by a Concerted Research Actions (GOA) grant from KU Leuven. S.J.J.B. is supported by NWO Vidi grant 864.11.005, European Research Council (ERC) Stg grant 639707 and a TU Delft start-up grant.
Author information
Authors and Affiliations
Contributions
F.L.N., L.L.D. and S.J.J.B. researched the data for the article. H.J.E.B., R.L., B.E.D. and S.J.J.B. provided substantial contribution to discussions of the content. F.L.N., M.V., P.A.J. and L.L.D. wrote the article. F.L.N., H.J.E.B., R.L., B.E.D. and S.J.J.B. reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Phages
-
Phages, or bacteriophages, are viruses that specifically infect bacteria for survival and replication.
- Porins
-
Proteins that assemble together to form a complex that acts as a channel (pore) across membranes through which molecules can diffuse.
- Baseplate
-
A protein structure found in tailed phages from the Myoviridae and Siphoviridae families. Tails and tail appendages, including tail fibres or tail spikes, attach to the baseplate.
- Prophages
-
Stable (relatively) forms of phage in which the genome is integrated into, and replicated with, the genome of the bacterial host, without lysis of the bacterial cell; prophages may also exist as extrachromosomal plasmids.
- O-antigens
-
Repetitive glycan polymers that constitute the outermost domain of the lipopolysaccharide of Gram-negative bacteria.
- Genetic plasticity
-
The alterable nature of genomes that enables the exchange of nucleic acids from one organism to another, usually to adapt to new environmental conditions.
- β-helix
-
A protein structure formed by parallel polypeptide chains associated in a helical pattern with either two or three faces; may be left-handed or right-handed.
- Vi capsule
-
A bacterial capsule that exposes the Vi antigen, a heat-labile somatic antigen thought to be associated with virulence in some bacteria.
- Deacetylation
-
The removal of an acetyl group from a chemical compound.
- Recombination
-
The process by which genetic material is exchanged between DNA molecules.
- Homotrimer
-
A protein composed of three identical units of polypeptide.
- Monomer
-
An individual molecule that can associate with similar molecules to form a larger molecule (dimer, trimer and polymer).
- Chaperones
-
Proteins that interact with and assist in the folding and unfolding or assembly and disassembly of other proteins without being part of the final structure.
- α-helix
-
A right-handed coiled conformation of proteins in which the resulting structure resembles a helix.
- Proteolysis
-
The breakdown (hydrolysis) of proteins or peptides into their key components, peptides and amino acids, by the action of enzymes.
- Phage tail-like complexes
-
Protein complexes that are homologous to phage tails and facilitate the killing of cells or influence bacterial and eukaryotic cells through physical interactions.
- Tropism
-
The orientation, by growth or movement, of all or part of an organism to an external stimulus.
Rights and permissions
About this article
Cite this article
Nobrega, F.L., Vlot, M., de Jonge, P.A. et al. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16, 760–773 (2018). https://doi.org/10.1038/s41579-018-0070-8
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-018-0070-8
This article is cited by
-
Characterization and genomics analysis of phage PGX1 against multidrug-resistant enterotoxigenic E. coli with in vivo and in vitro efficacy assessment
Animal Diseases (2024)
-
Structure of the intact tail machine of Anabaena myophage A-1(L)
Nature Communications (2024)
-
Application of BI-EHEC and BI-EPEC bacteriophages to control enterohemorrhagic and enteropathogenic escherichia coli on various food surfaces
BMC Research Notes (2023)
-
Variants of a putative baseplate wedge protein extend the host range of Pseudomonas phage K8
Microbiome (2023)
-
Mitigation of biogenic methanethiol using bacteriophages in synthetic wastewater augmented with Pseudomonas putida
Scientific Reports (2023)