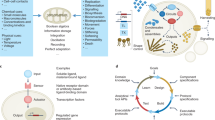
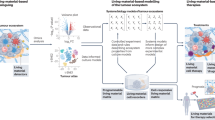
Abstract
Synthetic biology applies genetic tools to engineer living cells and organisms analogous to the programming of machines. In materials synthetic biology, engineering principles from synthetic biology and materials science are integrated to redesign living systems as dynamic and responsive materials with emerging and programmable functionalities. In this Review, we discuss synthetic-biology tools, including genetic circuits, model organisms and design parameters, which can be applied for the construction of smart living materials. We investigate non-living and living self-organizing multifunctional materials, such as intracellular structures and engineered biofilms, and examine the design and applications of hybrid living materials, including living sensors, therapeutics and electronics, as well as energy-conversion materials and living building materials. Finally, we consider prospects and challenges of programmable living materials and identify potential future applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sanchez, C., Arribart, H. & Giraud Guille, M. M. Biomimetism and bioinspiration as tools for the design of innovative materials and systems. Nat. Mater. 4, 277–288 (2005).
Liu, K. & Jiang, L. Bio-inspired design of multiscale structures for function integration. Nano Today 6, 155–175 (2011).
Wegst, U. G. K., Bai, H., Saiz, E., Tomsia, A. P. & Ritchie, R. O. Bioinspired structural materials. Nat. Mater. 14, 23–36 (2015).
Lu, Y., Aimetti, A. A., Langer, R. & Gu, Z. Bioresponsive materials. Nat. Rev. Mater. 2, 16075 (2016).
Palagi, S. & Fischer, P. Bioinspired microrobots. Nat. Rev. Mater. 3, 113–124 (2018).
Barthelat, F., Yin, Z. & Buehler, M. J. Structure and mechanics of interfaces in biological materials. Nat. Rev. Mater. 1, 16007 (2016).
Cameron, D. E., Bashor, C. J. & Collins, J. J. A brief history of synthetic biology. Nat. Rev. Microbiol. 12, 381–390 (2014).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Vecchio, D. D., Dy, A. J. & Qian, Y. Control theory meets synthetic biology. J. R. Soc. Interface 13, 20160380 (2016).
Seelig, G., Soloveichik, D., Zhang, D. Y. & Winfree, E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6, e16765 (2011).
Brophy, J. A. N. & Voigt, C. A. Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014).
Sedlmayer, F., Aubel, D. & Fussenegger, M. Synthetic gene circuits for the detection, elimination and prevention of disease. Nat. Biomed. Eng. 2, 399–415 (2018).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
Farzadfard, F. & Lu, T. K. Emerging applications for DNA writers and molecular recorders. Science 361, 870–875 (2018).
Ryu, M.-H. et al. Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol. 5, 314–330 (2020).
Praveschotinunt, P. et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 10, 5580 (2019).
Sun, G. L., Reynolds, E. E. & Belcher, A. M. Using yeast to sustainably remediate and extract heavy metals from waste waters. Nat. Sustain. 3, 303–311 (2020).
Heveran, C. M. et al. Biomineralization and successive regeneration of engineered living building materials. Matter 2, 481–494 (2020).
Smith, R. S. H. et al. Hybrid living materials: digital design and fabrication of 3D multimaterial structures with programmable biohybrid surfaces. Adv. Funct. Mater. 30, 1907401 (2020).
Chen, A. Y., Zhong, C. & Lu, T. K. Engineering living functional materials. ACS Synth. Biol. 4, 8–11 (2015).
Nguyen, P. Q., Courchesne, N.-M. D., Duraj-Thatte, A., Praveschotinunt, P. & Joshi, N. S. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 30, 1704847 (2018).
Basu, S., Gerchman, Y., Collins, C. H., Arnold, F. H. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005).
Chen, A. Y. et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523 (2014).
Toda, S., Blauch, L. R., Tang, S. K. Y., Morsut, L. & Lim, W. A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 361, 156–162 (2018).
DiMarco, R. L. & Heilshorn, S. C. Multifunctional materials through modular protein engineering. Adv. Mater. 24, 3923–3940 (2012).
Moradali, M. F. & Rehm, B. H. A. Bacterial biopolymers: from pathogenesis to advanced materials. Nat. Rev. Microbiol. 18, 195–210 (2020).
Rehm, B. H. A. Bacterial polymers: biosynthesis, modifications and applications. Nat. Rev. Microbiol. 8, 578–592 (2010).
Purnick, P. E. M. & Weiss, R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 10, 410–422 (2009).
Lee, S. Y. et al. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2, 18–33 (2019).
Lang, K. & Chin, J. W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 114, 4764–4806 (2014).
Elbaz, J., Yin, P. & Voigt, C. A. Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat. Commun. 7, 11179 (2016).
Wei, S.-P. et al. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol. 16, 1143–1148 (2020).
Meyer, A. J., Segall-Shapiro, T. H., Glassey, E., Zhang, J. & Voigt, C. A. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat. Chem. Biol. 15, 196–204 (2019).
Daniel, R., Rubens, J. R., Sarpeshkar, R. & Lu, T. K. Synthetic analog computation in living cells. Nature 497, 619–623 (2013).
Farzadfard, F. & Lu, T. K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014).
Levskaya, A. et al. Engineering Escherichia coli to see light. Nature 438, 441–442 (2005).
Piraner, D. I., Abedi, M. H., Moser, B. A., Lee-Gosselin, A. & Shapiro, M. G. Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol. 13, 75–80 (2017).
Ellis, T., Wang, X. & Collins, J. J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 27, 465–471 (2009).
Kelly, J. R. et al. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 3, 4 (2009).
Chen, Y.-J. et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659–664 (2013).
Tamsir, A., Tabor, J. J. & Voigt, C. A. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature 469, 212–215 (2011).
Wan, X. et al. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol. 15, 540–548 (2019).
Stricker, J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008).
Grindley, N. D. F., Whiteson, K. L. & Rice, P. A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 75, 567–605 (2006).
Bonnet, J., Subsoontorn, P. & Endy, D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl Acad. Sci. USA 109, 8884–8889 (2012).
Siuti, P., Yazbek, J. & Lu, T. K. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 31, 448–452 (2013).
Kalyoncu, E., Ahan, R. E., Ozcelik, C. E. & Seker, U. O. S. Genetic logic gates enable patterning of amyloid nanofibers. Adv. Mater. 31, 1902888 (2019).
Qi, Lei S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
McCarty, N. S., Graham, A. E., Studená, L. & Ledesma-Amaro, R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 11, 1281 (2020).
Gilbert, C. & Ellis, T. Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 8, 1–15 (2019).
Gao, X. J., Chong, L. S., Kim, M. S. & Elowitz, M. B. Programmable protein circuits in living cells. Science 361, 1252–1258 (2018).
Olson, E. J. & Tabor, J. J. Post-translational tools expand the scope of synthetic biology. Curr. Opin. Chem. Biol. 16, 300–306 (2012).
Green, et al. Toehold switches: de-novo-designed regulators of gene expression. Cell 159, 925–939 (2014).
Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R. & Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311 (2011).
Simon, A. J., d’Oelsnitz, S. & Ellington, A. D. Synthetic evolution. Nat. Biotechnol. 37, 730–743 (2019).
Rodriguez, E. A. et al. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129 (2017).
Thorne, N., Inglese, J. & Auld, D. S. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol. 17, 646–657 (2010).
Liljeruhm, J. et al. Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. J. Biol. Eng. 12, 8 (2018).
Narsing Rao, M. P., Xiao, M. & Li, W.-J. Fungal and bacterial pigments: secondary metabolites with wide applications. Front. Microbiol. 8, 1113 (2017).
Guo, Z., Richardson, J. J., Kong, B. & Liang, K. Nanobiohybrids: materials approaches for bioaugmentation. Sci. Adv. 6, eaaz0330 (2020).
Omenetto, F. G. & Kaplan, D. L. New opportunities for an ancient material. Science 329, 528–531 (2010).
Moon, T. S., Lou, C., Tamsir, A., Stanton, B. C. & Voigt, C. A. Genetic programs constructed from layered logic gates in single cells. Nature 491, 249–253 (2012).
Liu, Y. et al. Directing cellular information flow via CRISPR signal conductors. Nat. Methods 13, 938–944 (2016).
Roquet, N., Soleimany, A. P., Ferris, A. C., Aaronson, S. & Lu, T. K. Synthetic recombinase-based state machines in living cells. Science 353, aad8559 (2016).
Prindle, A. et al. A sensing array of radically coupled genetic ‘biopixels’. Nature 481, 39–44 (2012).
Billerbeck, S. et al. A scalable peptide-GPCR language for engineering multicellular communication. Nat. Commun. 9, 5057 (2018).
Zeng, J. et al. A synthetic microbial operational amplifier. ACS Synth. Biol. 7, 2007–2013 (2018).
Madsen, C. et al. Synthetic biology open language (SBOL) version 2.3. J. Integr. Bioinform. 16, 20190025 (2019).
Lee, K.-Y., Buldum, G., Mantalaris, A. & Bismarck, A. More than meets the eye in bacterial cellulose: biosynthesis, bioprocessing, and applications in advanced fiber composites. Macromol. Biosci. 14, 10–32 (2014).
Yadav, V. et al. Novel in vivo-degradable cellulose-chitin copolymer from metabolically engineered Gluconacetobacter xylinus. Appl. Environ. Microbiol. 76, 6257–6265 (2010).
Florea, M. et al. Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proc. Natl Acad. Sci. USA 113, E3431–E3440 (2016).
Abhijith, R., Ashok, A. & Rejeesh, C. R. Sustainable packaging applications from mycelium to substitute polystyrene: a review. Mater. Today Proc. 5, 2139–2145 (2018).
Wang, P.-A., Xiao, H. & Zhong, J.-J. CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum. Appl. Microbiol. Biotechnol. 104, 1661–1671 (2020).
Gilbert, C. et al. Living materials with programmable functionalities grown from engineered microbial co-cultures. Preprint at bioRxiv https://doi.org/10.1101/2019.12.20.882472 (2019)
Schaumberg, K. A. et al. Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat. Methods 13, 94–100 (2016).
Lienert, F., Lohmueller, J. J., Garg, A. & Silver, P. A. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 15, 95–107 (2014).
Mitiouchkina, T. et al. Plants with genetically encoded autoluminescence. Nat. Biotechnol. 38, 944–946 (2020).
Bredenoord, A. L., Clevers, H. & Knoblich, J. A. Human tissues in a dish: the research and ethical implications of organoid technology. Science 355, eaaf9414 (2017).
Kriegman, S., Blackiston, D., Levin, M. & Bongard, J. A scalable pipeline for designing reconfigurable organisms. Proc. Natl Acad. Sci. USA 117, 1853–1859 (2020).
Kassaw, T. K., Donayre-Torres, A. J., Antunes, M. S., Morey, K. J. & Medford, J. I. Engineering synthetic regulatory circuits in plants. Plant Sci. 273, 13–22 (2018).
Lew, T. T. S., Koman, V. B., Gordiichuk, P., Park, M. & Strano, M. S. The emergence of plant nanobionics and living plants as technology. Adv. Mater. Technol. 5, 1900657 (2020).
Franke, R. & Schreiber, L. Suberin — a biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol. 10, 252–259 (2007).
Li, F.-S., Phyo, P., Jacobowitz, J., Hong, M. & Weng, J.-K. The molecular structure of plant sporopollenin. Nat. Plants 5, 41–46 (2019).
Zhong, C. et al. Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat. Nanotechnol. 9, 858–866 (2014).
Nguyen, P. Q., Botyanszki, Z., Tay, P. K. R. & Joshi, N. S. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 5, 4945 (2014).
Huang, J. et al. Programmable and printable Bacillus subtilis biofilms as engineered living materials. Nat. Chem. Biol. 15, 34–41 (2019).
Bourdeau, R. W. et al. Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature 553, 86–90 (2018).
Tay, P. K. R., Nguyen, P. Q. & Joshi, N. S. A synthetic circuit for mercury bioremediation using self-assembling functional amyloids. ACS Synth. Biol. 6, 1841–1850 (2017).
Zhang, C. et al. Engineered Bacillus subtilis biofilms as living glues. Mater. Today 28, 40–48 (2019).
Liu, X. et al. 3D printing of living responsive materials and devices. Adv. Mater. 30, 1704821 (2018).
Tang, T.-C. et al. Tough hydrogel-based biocontainment of engineered organisms for continuous, self-powered sensing and computation. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2020.02.11.941120v1 (2020).
Whitesides, G. M. & Grzybowski, B. Self-assembly at all scales. Science 295, 2418–2421 (2002).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Dong, Y. et al. DNA functional materials assembled from branched DNA: design, synthesis, and applications. Chem. Rev. 120, 9420–9481 (2020).
Woolston, B. M., Edgar, S. & Stephanopoulos, G. Metabolic engineering: past and future. Annu. Rev. Chem. Biomol. Eng. 4, 259–288 (2013).
Wagner, H. J. et al. Synthetic biology-inspired design of signal-amplifying materials systems. Mater. Today 22, 25–34 (2019).
Pena-Francesch, A., Jung, H., Demirel, M. C. & Sitti, M. Biosynthetic self-healing materials for soft machines. Nat. Mater. 19, 1230–1235 (2020).
English, M. A. et al. Programmable CRISPR-responsive smart materials. Science 365, 780–785 (2019).
Cui, M. et al. Exploiting mammalian low-complexity domains for liquid-liquid phase separation–driven underwater adhesive coatings. Sci. Adv. 5, eaax3155 (2019).
Wallace, A. K., Chanut, N. & Voigt, C. A. Silica nanostructures produced using diatom peptides with designed post-translational modifications. Adv. Funct. Mater. 23, 2000849 (2020).
Amiram, M. et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol. 33, 1272–1279 (2015).
Qian, Z.-G., Pan, F. & Xia, X.-X. Synthetic biology for protein-based materials. Curr. Opin. Biotechnol. 65, 197–204 (2020).
Keating, K. W. & Young, E. M. Synthetic biology for bio-derived structural materials. Curr. Opin. Chem. Eng. 24, 107–114 (2019).
Meng, D.-C. et al. Production and characterization of poly(3-hydroxypropionate-co-4-hydroxybutyrate) with fully controllable structures by recombinant Escherichia coli containing an engineered pathway. Metab. Eng. 14, 317–324 (2012).
Deepankumar, K. et al. Supramolecular β-sheet suckerin–based underwater adhesives. Adv. Funct. Mater. 30, 1907534 (2020).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Bracha, D., Walls, M. T. & Brangwynne, C. P. Probing and engineering liquid-phase organelles. Nat. Biotechnol. 37, 1435–1445 (2019).
Nakamura, H. et al. Intracellular production of hydrogels and synthetic RNA granules by multivalent molecular interactions. Nat. Mater. 17, 79–89 (2018).
Kolinko, I. et al. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat. Nanotechnol. 9, 193–197 (2014).
Chatterjee, A. et al. Cephalopod-inspired optical engineering of human cells. Nat. Commun. 11, 2708 (2020).
Farhadi, A., Ho, G. H., Sawyer, D. P., Bourdeau, R. W. & Shapiro, M. G. Ultrasound imaging of gene expression in mammalian cells. Science 365, 1469–1475 (2019).
Flemming, H.-C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
Rumbaugh, K. P. & Sauer, K. Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586 (2020).
Knowles, T. P. J. & Buehler, M. J. Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 6, 469–479 (2011).
Blanco, L. P., Evans, M. L., Smith, D. R., Badtke, M. P. & Chapman, M. R. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20, 66–73 (2012).
Barnhart, M. M. & Chapman, M. R. Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 (2006).
Wang, X. et al. Programming cells for dynamic assembly of inorganic nano-objects with spatiotemporal control. Adv. Mater. 30, 1705968 (2018).
Kalyoncu, E., Ahan, R. E., Olmez, T. T. & Safak Seker, U. O. Genetically encoded conductive protein nanofibers secreted by engineered cells. RSC Adv. 7, 32543–32551 (2017).
Dorval Courchesne, N.-M. et al. Biomimetic engineering of conductive curli protein films. Nanotechnology 29, 454002 (2018).
Jiang, L. et al. Programming integrative extracellular and intracellular biocatalysis for rapid, robust, and recyclable synthesis of trehalose. ACS Catal. 8, 1837–1842 (2018).
Botyanszki, Z., Tay, P. K. R., Nguyen, P. Q., Nussbaumer, M. G. & Joshi, N. S. Engineered catalytic biofilms: Site-specific enzyme immobilization onto E. coli curli nanofibers. Biotechnol. Bioeng. 112, 2016–2024 (2015).
Pu, J. et al. Virus disinfection from environmental water sources using living engineered biofilm materials. Adv. Sci. 7, 1903558 (2020).
Wang, X. et al. Immobilization of functional nano-objects in living engineered bacterial biofilms for catalytic applications. Natl Sci. Rev. 6, 929–943 (2019).
Seker, U. O. S., Chen, A. Y., Citorik, R. J. & Lu, T. K. Synthetic biogenesis of bacterial amyloid nanomaterials with tunable inorganic–organic interfaces and electrical conductivity. ACS Synth. Biol. 6, 266–275 (2017).
An, B. et al. Programming living glue systems to perform autonomous mechanical repairs. Matter https://doi.org/10.1016/j.matt.2020.09.006 (2020).
Charrier, M. et al. Engineering the S-layer of Caulobacter crescentus as a foundation for stable, high-density, 2D living materials. ACS Synth. Biol. 8, 181–190 (2019).
Fang, J., Kawano, S., Tajima, K. & Kondo, T. In vivo curdlan/cellulose bionanocomposite synthesis by genetically modified Gluconacetobacter xylinus. Biomacromolecules 16, 3154–3160 (2015).
Walker, K. T., Goosens, V. J., Das, A., Graham, A. E. & Ellis, T. Engineered cell-to-cell signalling within growing bacterial cellulose pellicles. Microb. Biotechnol. 12, 611–619 (2019).
Fan, G., Graham, A. J., Kolli, J., Lynd, N. A. & Keitz, B. K. Aerobic radical polymerization mediated by microbial metabolism. Nat. Chem. 12, 638–646 (2020).
Fan, G., Dundas, C. M., Graham, A. J., Lynd, N. A. & Keitz, B. K. Shewanella oneidensis as a living electrode for controlled radical polymerization. Proc. Natl Acad. Sci. USA 115, 4559–4564 (2018).
Gao, M. et al. A natural in situ fabrication method of functional bacterial cellulose using a microorganism. Nat. Commun. 10, 437 (2019).
Koch, A. J. & Meinhardt, H. Biological pattern formation: from basic mechanisms to complex structures. Rev. Mod. Phys. 66, 1481–1507 (1994).
Salazar-Ciudad, I., Jernvall, J. & Newman, S. A. Mechanisms of pattern formation in development and evolution. Development 130, 2027–2037 (2003).
Kondo, S. & Miura, T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620 (2010).
Luo, N., Wang, S. & You, L. Synthetic pattern formation. Biochemistry 58, 1478–1483 (2019).
Kim, H., Jin, X., Glass, D. S. & Riedel-Kruse, I. H. Engineering and modeling of multicellular morphologies and patterns. Curr. Opin. Genet. Dev. 63, 95–102 (2020).
Santos-Moreno, J. & Schaerli, Y. Using synthetic biology to engineer spatial patterns. Adv. Biosyst. 3, 1800280 (2019).
Fernandez-Rodriguez, J., Moser, F., Song, M. & Voigt, C. A. Engineering RGB color vision into Escherichia coli. Nat. Chem. Biol. 13, 706–708 (2017).
Moser, F., Tham, E., González, L. M., Lu, T. K. & Voigt, C. A. Light-controlled, high-resolution patterning of living engineered bacteria onto textiles, ceramics, and plastic. Adv. Funct. Mater. 29, 1901788 (2019).
Liu, C. et al. Sequential establishment of stripe patterns in an expanding cell population. Science 334, 238–241 (2011).
Tabor, J. J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009).
Turing, A. M. The chemical basis of morphogenesis. Bull. Math. Biol. 52, 153–197 (1990).
Karig, D. et al. Stochastic Turing patterns in a synthetic bacterial population. Proc. Natl Acad. Sci. USA 115, 6572–6577 (2018).
Potvin-Trottier, L., Lord, N. D., Vinnicombe, G. & Paulsson, J. Synchronous long-term oscillations in a synthetic gene circuit. Nature 538, 514–517 (2016).
Mushnikov, N. V., Fomicheva, A., Gomelsky, M. & Bowman, G. R. Inducible asymmetric cell division and cell differentiation in a bacterium. Nat. Chem. Biol. 15, 925–931 (2019).
Molinari, S. et al. A synthetic system for asymmetric cell division in Escherichia coli. Nat. Chem. Biol. 15, 917–924 (2019).
Glass, D. S. & Riedel-Kruse, I. H. A synthetic bacterial cell-cell adhesion toolbox for programming multicellular morphologies and patterns. Cell 174, 649–658.e16 (2018).
Perry, C. C. & Keeling-Tucker, T. Biosilicification: the role of the organic matrix in structure control. J. Biol. Inorg. Chem. 5, 537–550 (2000).
van der Meer, J. R. & Belkin, S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat. Rev. Microbiol. 8, 511–522 (2010).
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Li, S., Li, Y. & Smolke, C. D. Strategies for microbial synthesis of high-value phytochemicals. Nat. Chem. 10, 395–404 (2018).
Bereza-Malcolm, L. T., Mann, G. & Franks, A. E. Environmental sensing of heavy metals through whole cell microbial biosensors: a synthetic biology approach. ACS Synth. Biol. 4, 535–546 (2015).
Ostrov, N. et al. A modular yeast biosensor for low-cost point-of-care pathogen detection. Sci. Adv. 3, e1603221 (2017).
Belkin, S. et al. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 35, 308–310 (2017).
Liu, X. et al. Stretchable living materials and devices with hydrogel–elastomer hybrids hosting programmed cells. Proc. Natl Acad. Sci. USA 114, 2200–2205 (2017).
Landry, B. P., Palanki, R., Dyulgyarov, N., Hartsough, L. A. & Tabor, J. J. Phosphatase activity tunes two-component system sensor detection threshold. Nat. Commun. 9, 1433 (2018).
Chen, Y. et al. Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 64 (2018).
Salis, H. M., Mirsky, E. A. & Voigt, C. A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950 (2009).
Shaw, W. M. et al. Engineering a model cell for rational tuning of GPCR signaling. Cell 177, 782–796.e27 (2019).
Maxmen, A. Living therapeutics: Scientists genetically modify bacteria to deliver drugs. Nat. Med. 23, 5–7 (2017).
Bose, S. et al. A retrievable implant for the long-term encapsulation and survival of therapeutic xenogeneic cells. Nat. Biomed. Eng. 4, 814–826 (2020).
Sankaran, S. & del Campo, A. Optoregulated protein release from an engineered living material. Adv. Biosyst. 3, 1800312 (2019).
Sankaran, S., Becker, J., Wittmann, C. & del Campo, A. Optoregulated drug release from an engineered living material: self-replenishing drug depots for long-term, light-regulated delivery. Small 15, 1804717 (2019).
Dai, Z. et al. Versatile biomanufacturing through stimulus-responsive cell–material feedback. Nat. Chem. Biol. 15, 1017–1024 (2019).
Gerber, L. C., Koehler, F. M., Grass, R. N. & Stark, W. J. Incorporation of penicillin-producing fungi into living materials to provide chemically active and antibiotic-releasing surfaces. Angew. Chem. Int. Ed. 124, 11455–11458 (2012).
González, L. M., Mukhitov, N. & Voigt, C. A. Resilient living materials built by printing bacterial spores. Nat. Chem. Biol. 16, 126–133 (2020).
Sankaran, S., Zhao, S., Muth, C., Paez, J. & del Campo, A. Toward light-regulated living biomaterials. Adv. Sci. 5, 1800383 (2018).
Saadeddin, A. et al. Functional living biointerphases. Adv. Healthc. Mater. 2, 1213–1218 (2013).
Hay, J. J. et al. Living biointerfaces based on non-pathogenic bacteria support stem cell differentiation. Sci. Rep. 6, 21809 (2016).
Hay, J. J. et al. Bacteria-based materials for stem cell engineering. Adv. Mater. 30, 1804310 (2018).
Rodrigo-Navarro, A., Rico, P., Saadeddin, A., Garcia, A. J. & Salmeron-Sanchez, M. Living biointerfaces based on non-pathogenic bacteria to direct cell differentiation. Sci. Rep. 4, 5849 (2014).
Lufton, M. et al. Living bacteria in thermoresponsive gel for treating fungal infections. Adv. Funct. Mater. 28, 1801581 (2018).
Mimee, M. et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918 (2018).
Din, M. O., Martin, A., Razinkov, I., Csicsery, N. & Hasty, J. Interfacing gene circuits with microelectronics through engineered population dynamics. Sci. Adv. 6, eaaz8344 (2020).
Patel, S. R. & Lieber, C. M. Precision electronic medicine in the brain. Nat. Biotechnol. 37, 1007–1012 (2019).
Webster, D. P. et al. An arsenic-specific biosensor with genetically engineered Shewanella oneidensis in a bioelectrochemical system. Biosens. Bioelectron. 62, 320–324 (2014).
Shao, J. et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice. Sci. Transl. Med. 9, eaal2298 (2017).
Tschirhart, T. et al. Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nat. Commun. 8, 14030 (2017).
Krawczyk, K. et al. Electrogenetic cellular insulin release for real-time glycemic control in type 1 diabetic mice. Science 368, 993–1001 (2020).
Slate, A. J., Whitehead, K. A., Brownson, D. A. & Banks, C. E. Microbial fuel cells: An overview of current technology. Renew. Sustain. Energy Rev. 101, 60–81 (2019).
Bird, L. J. et al. Engineered living conductive biofilms as functional materials. MRS Commun. 9, 505–517 (2019).
Li, F., Wang, L., Liu, C., Wu, D. & Song, H. Engineering exoelectrogens by synthetic biology strategies. Curr. Opin. Electrochem. 10, 37–45 (2018).
Gadhamshetty, V. & Koratkar, N. Nano-engineered biocatalyst-electrode structures for next generation microbial fuel cells. Nano Energy 1, 3–5 (2012).
Yong, Y.-C., Yu, Y.-Y., Zhang, X. & Song, H. Highly active bidirectional electron transfer by a self-assembled electroactive reduced-graphene-oxide-hybridized biofilm. Angew. Chem. Int. Ed. 53, 4480–4483 (2014).
McCormick, A. J. et al. Photosynthetic biofilms in pure culture harness solar energy in a mediatorless bio-photovoltaic cell (BPV) system. Energy Environ. Sci. 4, 4699–4709 (2011).
Joshi, S., Cook, E. & Mannoor, M. S. Bacterial nanobionics via 3D printing. Nano Lett. 18, 7448–7456 (2018).
Melis, A. Solar energy conversion efficiencies in photosynthesis: minimizing the chlorophyll antennae to maximize efficiency. Plant Sci. 177, 272–280 (2009).
Kim, M. J. et al. A broadband multiplex living solar cell. Nano Lett. 20, 4286–4291 (2020).
Schuergers, N., Werlang, C., Ajo-Franklin, C. M. & Boghossian, A. A. A synthetic biology approach to engineering living photovoltaics. Energy Environ. Sci. 10, 1102–1115 (2017).
Cestellos-Blanco, S., Zhang, H., Kim, J. M., Shen, Y.-X. & Yang, P. Photosynthetic semiconductor biohybrids for solar-driven biocatalysis. Nat. Catal. 3, 245–255 (2020).
Sakimoto, K. K., Wong, A. B. & Yang, P. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science 351, 74–77 (2016).
Wei, W. et al. A surface-display biohybrid approach to light-driven hydrogen production in air. Sci. Adv. 4, eaap9253 (2018).
Guo, J. et al. Light-driven fine chemical production in yeast biohybrids. Science 362, 813–816 (2018).
Bernardi, D., DeJong, J. T., Montoya, B. M. & Martinez, B. C. Bio-bricks: Biologically cemented sandstone bricks. Constr. Build. Mater. 55, 462–469 (2014).
Lee, Y. S. & Park, W. Current challenges and future directions for bacterial self-healing concrete. Appl. Microbiol. Biotechnol. 102, 3059–3070 (2018).
Pungrasmi, W., Intarasoontron, J., Jongvivatsakul, P. & Likitlersuang, S. Evaluation of microencapsulation techniques for MICP bacterial spores applied in self-healing concrete. Sci. Rep. 9, 12484 (2019).
Boothby, T. C. et al. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol. Cell 65, 975–984.e975 (2017).
Ferreiro, A., Crook, N., Gasparrini, A. J. & Dantas, G. Multiscale evolutionary dynamics of host-associated microbiomes. Cell 172, 1216–1227 (2018).
Jones, M., Huynh, T., Dekiwadia, C., Daver, F. & John, S. Mycelium composites: a review of engineering characteristics and growth kinetics. J. Bionanosci. 11, 241–257 (2017).
Chang, J. et al. Modified recipe to inhibit fruiting body formation for living fungal biomaterial manufacture. PLoS ONE 14, e0209812 (2019).
Islam, M. R., Tudryn, G., Bucinell, R., Schadler, L. & Picu, R. C. Mechanical behavior of mycelium-based particulate composites. J. Mater. Sci. 53, 16371–16382 (2018).
Jiang, B. et al. Lignin as a wood-inspired binder enabled strong, water stable, and biodegradable paper for plastic replacement. Adv. Funct. Mater. 30, 1906307 (2020).
Teulé, F. et al. Silkworms transformed with chimeric silkworm/spider silk genes spin composite silk fibers with improved mechanical properties. Proc. Natl Acad. Sci. USA 109, 923–928 (2012).
Tero, A. et al. Rules for biologically inspired adaptive network design. Science 327, 439–442 (2010).
Inda, M. E. & Lu, T. K. Microbes as biosensors. Annu. Rev. Microbiol. 74, 337–359 (2020).
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
Morrison, M. S., Podracky, C. J. & Liu, D. R. The developing toolkit of continuous directed evolution. Nat. Chem. Biol. 16, 610–619 (2020).
Gleizer, S. et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 179, 1255–1263.e12 (2019).
Hossain, A. et al. Automated design of thousands of nonrepetitive parts for engineering stable genetic systems. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0584-2 (2020).
Nielsen, A. A. K. et al. Genetic circuit design automation. Science 352, aac7341 (2016).
Casini, A., Storch, M., Baldwin, G. S. & Ellis, T. Bricks and blueprints: methods and standards for DNA assembly. Nat. Rev. Mol. Cell Biol. 16, 568–576 (2015).
Zhang, W., Mitchell, L. A., Bader, J. S. & Boeke, J. D. Synthetic genomes. Annu. Rev. Biochem. 89, 77–101 (2020).
Adamala, K. P., Martin-Alarcon, D. A., Guthrie-Honea, K. R. & Boyden, E. S. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 9, 431–439 (2017).
Ceroni, F. et al. Burden-driven feedback control of gene expression. Nat. Methods 15, 387–393 (2018).
Segall-Shapiro, T. H., Meyer, A. J., Ellington, A. D., Sontag, E. D. & Voigt, C. A. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol. 10, 742 (2014).
Burger, B. et al. A mobile robotic chemist. Nature 583, 237–241 (2020).
Butler, K. T., Davies, D. W., Cartwright, H., Isayev, O. & Walsh, A. Machine learning for molecular and materials science. Nature 559, 547–555 (2018).
Stokes, J. M. et al. A deep learning approach to antibiotic discovery. Cell 180, 688–702.e13 (2020).
Camacho, D. M., Collins, K. M., Powers, R. K., Costello, J. C. & Collins, J. J. Next-generation machine learning for biological networks. Cell 173, 1581–1592 (2018).
Qin, Z. et al. Artificial intelligence method to design and fold alpha-helical structural proteins from the primary amino acid sequence. Extreme Mech. Lett. 36, 100652 (2020).
Wong, B. G., Mancuso, C. P., Kiriakov, S., Bashor, C. J. & Khalil, A. S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 36, 614–623 (2018).
Lee, J. W., Chan, C. T., Slomovic, S. & Collins, J. J. Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol. 14, 530–537 (2018).
Rovner, A. J. et al. Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93 (2015).
McLeod, C. & Nerlich, B. Synthetic biology, metaphors and responsibility. Life Sci. Soc. Policy 13, 13 (2017).
Trump, B. D. et al. Co-evolution of physical and social sciences in synthetic biology. Crit. Rev. Biotechnol. 39, 351–365 (2019).
Levin, M., Bongard, J. & Lunshof, J. E. Applications and ethics of computer-designed organisms. Nat. Rev. Mol. Cell Biol. 21, 655–656 (2020).
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997).
Zuo, J., Niu, Q.-W. & Chua, N.-H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 (2000).
Motta-Mena, L. B. et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 10, 196–202 (2014).
Inda, M. E., Vazquez, D. B., Fernández, A. & Cybulski, L. E. Reverse engineering of a thermosensing regulator switch. J. Mol. Biol. 431, 1016–1024 (2019).
Booth, I. R., Edwards, M. D., Black, S., Schumann, U. & Miller, S. Mechanosensitive channels in bacteria: signs of closure? Nat. Rev. Microbiol. 5, 431–440 (2007).
Callura, J. M., Dwyer, D. J., Isaacs, F. J., Cantor, C. R. & Collins, J. J. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl Acad. Sci. USA 107, 15898–15903 (2010).
Rhodius, V. A. et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 9, 702 (2013).
Gander, M. W., Vrana, J. D., Voje, W. E., Carothers, J. M. & Klavins, E. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat. Commun. 8, 15459 (2017).
Sheth, R. U., Yim, S. S., Wu, F. L. & Wang, H. H. Multiplex recording of cellular events over time on CRISPR biological tape. Science 358, 1457–1461 (2017).
Friedland, A. E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009).
Tastanova, A. et al. Synthetic biology-based cellular biomedical tattoo for detection of hypercalcemia associated with cancer. Sci. Transl. Med. 10, eaap8562 (2018).
Chen, G.-Q., Jiang, X.-R. & Guo, Y. Synthetic biology of microbes synthesizing polyhydroxyalkanoates (PHA). Synth. Syst. Biotechnol. 1, 236–242 (2016).
Jensen, H. M. et al. Engineering of a synthetic electron conduit in living cells. Proc. Natl Acad. Sci. USA 107, 19213–19218 (2010).
Piñero-Lambea, C. et al. Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth. Biol. 4, 463–473 (2015).
Teramoto, H. et al. Genetic code expansion of the silkworm Bombyx mori to functionalize silk fiber. ACS Synth. Biol. 7, 801–806 (2018).
Sun, F., Zhang, W.-B., Mahdavi, A., Arnold, F. H. & Tirrell, D. A. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc. Natl Acad. Sci. USA 111, 11269–11274 (2014).
Deng, M.-D. et al. Metabolic engineering of Escherichia coli for industrial production of glucosamine and N-acetylglucosamine. Metab. Eng. 7, 201–214 (2005).
Nishida, K. & Silver, P. A. Induction of biogenic magnetization and redox control by a component of the target of rapamycin complex 1 signaling pathway. PLoS Biol. 10, e1001269 (2012).
Liu, X. et al. Engineering genetically-encoded mineralization and magnetism via directed evolution. Sci. Rep. 6, 38019 (2016).
Liang, L. et al. Rational control of calcium carbonate precipitation by engineered Escherichia coli. ACS Synth. Biol. 7, 2497–2506 (2018).
Cui, R. et al. Living yeast cells as a controllable biosynthesizer for fluorescent quantum dots. Adv. Funct. Mater. 19, 2359–2364 (2009).
Rivera-Tarazona, L. K., Bhat, V. D., Kim, H., Campbell, Z. T. & Ware, T. H. Shape-morphing living composites. Sci. Adv. 6, eaax8582 (2020).
Schaffner, M., Rühs, P. A., Coulter, F., Kilcher, S. & Studart, A. R. 3D printing of bacteria into functional complex materials. Sci. Adv. 3, eaao6804 (2017).
Tang, J. et al. Cardiac cell–integrated microneedle patch for treating myocardial infarction. Sci. Adv. 4, eaat9365 (2018).
Ye, H. et al. Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat. Biomed. Eng. 1, 0005 (2016).
An, D. et al. Designing a retrievable and scalable cell encapsulation device for potential treatment of type 1 diabetes. Proc. Natl Acad. Sci. USA 115, E263–E272 (2018).
Guo, S. et al. Engineered living materials based on adhesin-mediated trapping of programmable cells. ACS Synth. Biol. 9, 475–485 (2020).
Fu, T.-M., Hong, G., Viveros, R. D., Zhou, T. & Lieber, C. M. Highly scalable multichannel mesh electronics for stable chronic brain electrophysiology. Proc. Natl Acad. Sci. USA 114, E10046–E10055 (2017).
Cao, Y. et al. Programmable assembly of pressure sensors using pattern-forming bacteria. Nat. Biotechnol. 35, 1087–1093 (2017).
McCuskey, S. R., Su, Y., Leifert, D., Moreland, A. S. & Bazan, G. C. Living bioelectrochemical composites. Adv. Mater. 32, 1908178 (2020).
Freyman, M. C., Kou, T., Wang, S. & Li, Y. 3D printing of living bacteria electrode. Nano Res. 13, 1318–1323 (2020).
Liu, C. et al. Nanowire–bacteria hybrids for unassisted solar carbon dioxide fixation to value-added chemicals. Nano Lett. 15, 3634–3639 (2015).
Zhang, H. et al. Bacteria photosensitized by intracellular gold nanoclusters for solar fuel production. Nat. Nanotechnol. 13, 900–905 (2018).
Honda, Y., Hagiwara, H., Ida, S. & Ishihara, T. Application to photocatalytic H2 production of a whole-cell reaction by recombinant Escherichia coli cells expressing [FeFe]-hydrogenase and maturases genes. Angew. Chem. Int. Ed. 55, 8045–8048 (2016).
Sun, W., Tajvidi, M., Hunt, C. G., McIntyre, G. & Gardner, D. J. Fully bio-based hybrid composites made of wood, fungal mycelium and cellulose nanofibrils. Sci. Rep. 9, 3766 (2019).
Wood, T. L. et al. Living biofouling-resistant membranes as a model for the beneficial use of engineered biofilms. Proc. Natl Acad. Sci. USA 113, E2802–E2811 (2016).
Johnston, T. G. et al. Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation. Nat. Commun. 11, 563 (2020).
Qian, F. et al. Direct writing of tunable living inks for bioprocess intensification. Nano Lett. 19, 5829–5835 (2019).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Lee, S.-W., Mao, C., Flynn, C. E. & Belcher, A. M. Ordering of quantum dots using genetically engineered viruses. Science 296, 892–895 (2002).
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Annaluru, N. et al. Total synthesis of a functional designer eukaryotic chromosome. Science 344, 55–58 (2014).
Ye, H. & Fussenegger, M. Synthetic therapeutic gene circuits in mammalian cells. FEBS Lett. 588, 2537–2544 (2014).
Chen, Z. et al. De novo design of protein logic gates. Science 368, 78–84 (2020).
Acknowledgements
The authors acknowledge Mr. Lei Chen for assistance in preparing the figures and Dr. Karen Pepper for reviewing the manuscript. B.A. would like to thank the support provided by the China Scholarship Council (CSC) during his visiting period at Massachusetts Institute of Technology. This work was sponsored by the National Key R&D Program of China (grant nos. 2020YFA0908100 and 2018YFA0902804, the two grants provide equal support), the Joint Funds of the National Natural Science Foundation of China (key program no. U1932204), the National Institutes of Health of the USA (grant no. 1-R21-AI121669-01) and the Defense Threat Reduction Agency of the USA (grant no. HDTRA1-15-1-0050).
Author information
Authors and Affiliations
Contributions
All authors contributed to the discussions, writing and reviewing of the article content. T.-C.T., B.A. and C.Z. prepared the figures and tables.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tang, TC., An, B., Huang, Y. et al. Materials design by synthetic biology. Nat Rev Mater 6, 332–350 (2021). https://doi.org/10.1038/s41578-020-00265-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41578-020-00265-w
This article is cited by
-
Self-pigmenting textiles grown from cellulose-producing bacteria with engineered tyrosinase expression
Nature Biotechnology (2024)
-
Accelerating the design of pili-enabled living materials using an integrative technological workflow
Nature Chemical Biology (2024)
-
Synthetic microbiology in sustainability applications
Nature Reviews Microbiology (2024)
-
Genetically targeted chemical assembly
Nature Reviews Bioengineering (2023)
-
Synthetic living materials in cancer biology
Nature Reviews Bioengineering (2023)