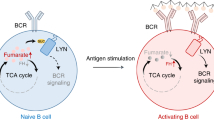
Abstract
B cells face multiple restrictions on glucose and energy metabolism. Their lineage-determining transcription factors repress glucose uptake and pentose phosphate pathway activity, while their low numbers of mitochondria and small cytoplasmic volume set narrow limits for mitochondrial ATP production and autophagy as alternative energy sources. During activation, B cells can balance temporary increases of energy expenditure. However, permanent hyperactivation of kinases, for instance, downstream of an autoreactive B cell receptor (BCR) or a transforming oncogene, can cause energy stress and cell death. Here, I propose that B cell-intrinsic restriction of ATP represents a safeguard to eliminate autoreactive or pre-malignant B cells. If the metabolic gatekeepers are compromised, influx of additional glucose may fuel permanent increases in metabolic demands and pathological B cell proliferation, driven by an autoreactive BCR or a transforming oncogene.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rajewsky, K. Clonal selection and learning in the antibody system. Nature 381, 751–758 (1996).
Klein, F. et al. Tracing the pre-B to immature B cell transition in human leukemia cells reveals a coordinated sequence of primary and secondary IGK gene rearrangement, IGK deletion, and IGL gene rearrangement. J. Immunol. 174, 367–375 (2005).
Schatz, D. G., Oettinger, M. A. & Baltimore, D. The V(D)J recombination activating gene (RAG-1). Cell 59, 1035–1048 (1989).
Kitagawa, Y. et al. Prevalent involvement of illegitimate V(D)J recombination in chromosome 9p21 deletions in lymphoid leukemia. J. Biol. Chem. 277, 46289–46297 (2002).
Tsai, A. G. et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell 135, 1130–1142 (2008).
Papaemmanuil, E. et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat. Genet. 46, 116–125 (2014).
Swaminathan, S. et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat. Immunol. 16, 766–774 (2015).
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2001).
Ramiro, A. R. et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118, 431–438 (2004).
Pasqualucci, L. et al. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40, 108–112 (2008).
Klemm, L. et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell 16, 232–245 (2009).
Jankovic, M. et al. 53BP1 alters the landscape of DNA rearrangements and suppresses AID-induced B cell lymphoma. Mol. Cell 49, 623–631 (2013).
Compagno, M. et al. Phosphatidylinositol 3-kinase δ blockade increases genomic instability in B cells. Nature 542, 489–493 (2017).
Cazzaniga, G. et al. Developmental origins and impact of BCR-ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood 118, 5559–5564 (2011).
Gale, K. B. et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc. Natl Acad. Sci. USA 94, 13950–13954 (1997).
Wiemels, J. L. et al. Prenatal origin of acute lymphoblastic leukemia in children. Lancet 354, 1499–1503 (1999).
Biernaux, C. et al. Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood 86, 3118–3122 (1995).
Bose, S. et al. The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood 92, 3362–3367 (1998).
Takagi, M. et al. Autoimmune lymphoproliferative syndrome-like disease with somatic KRAS mutation. Blood 117, 2887–2890 (2011).
Osmond, D. G. Proliferation kinetics and the lifespan of B cells in central and peripheral lymphoid organs. Curr. Opin. Immunol. 3, 179–185 (1991).
Wardemann, H. et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377 (2003).
Hardy, R. R. & Hayakawa, K. B cell development pathways. Annu. Rev. Immunol. 19, 595–621 (2001).
Waters, L. R. et al. Initial B cell activation induces metabolic reprogramming and mitochondrial remodeling. iScience 5, 99–109 (2018).
Dufort, F. J. et al. Cutting edge: IL-4-mediated protection of primary B lymphocytes from apoptosis via Stat6-dependent regulation of glycolytic metabolism. J. Immunol. 179, 4953–4957 (2007).
Cho, S. H. et al. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature 537, 234–238 (2016).
Caro-Maldonado, A. et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J. Immunol. 192, 3626–3636 (2014).
Akkaya, M. et al. Second signals rescue B cells from activation-induced mitochondrial dysfunction and death. Nat. Immunol. 19, 871–884 (2018).
Liu, T. et al. Glucose transporter 1-mediated glucose uptake is limiting for B cell acute lymphoblastic leukemia anabolic metabolism and resistance to apoptosis. Cell Death Dis. 5, e1470 (2014).
Chan, L. N. et al. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 542, 479–483 (2017).
Jiang, S. et al. Let-7 suppresses B cell activation through restricting the availability of necessary nutrients. Cell Metab. 27, 393–403 (2018).
Tiegs, S. L., Russell, D. M. & Nemazee, D. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177, 1009–1020 (1993).
Shojaee, S. et al. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat. Med. 22, 379–387 (2016).
Chen, Z. et al. Signalling thresholds and negative B cell selection in acute lymphoblastic leukemia. Nature 521, 357–361 (2015).
Chan, L. N. & Müschen, M. B cell identity as a metabolic barrier against malignant transformation. Exp. Hematol. 53, 1–6 (2017).
Martín-Lorenzo, A. Loss of Pax5 exploits Sca1-BCR-ABLp190 susceptibility to confer the metabolic shift essential for pre-B ALL. Cancer Res. 78, 2669–2679 (2018).
McFadden, K. et al. Metabolic stress is a barrier to Epstein-Barr virus-mediated B cell immortalization. Proc. Natl Acad. Sci. USA 113, E782–E790 (2016).
Bhatt, A. P. et al. Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc. Natl Acad. Sci. USA 109, 11818–11823 (2012).
Lu, Z. et al. Fasting selectively blocks development of acute lymphoblastic leukemia via leptin-receptor upregulation. Nat. Med. 23, 79–90 (2017).
Xiao, G. et al. B-cell-specific diversion of glucose carbon utilization reveals a unique vulnerability in B cell malignancies. Cell 173, 470–484 (2018).
Müschen, M. Autoimmunity checkpoints as therapeutic targets in B cell malignancies. Nat. Rev. Cancer 18, 103–116 (2018).
Zhang, C. S. et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 (2017).
Wu, N. et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol. Cell 49, 1167–1175 (2013).
Xie, H. et al. Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676 (2004).
Faubert, B. et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 17, 113–124 (2013).
Butturini, A. M. et al. Obesity and outcome in pediatric acute lymphoblastic leukemia. J. Clin. Oncol. 25, 2063–2069 (2007).
Gelelete, C. B. et al. Overweight as a prognostic factor in children with acute lymphoblastic leukemia. Obesity 19, 1908–1911 (2011).
Orgel, E. et al. Obesity is associated with residual leukemia following induction therapy for childhood B-precursor acute lymphoblastic leukemia. Blood 124, 3932–3938 (2014).
Weiser, M. A. et al. Relation between the duration of remission and hyperglycemia during induction chemotherapy for acute lymphocytic leukemia with a hyperfractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone/methotrexate-cytarabine regimen. Cancer 100, 1179–1185 (2004).
Castillo, J. J. et al. Obesity is associated with increased relative risk of diffuse large B cell lymphoma: a meta-analysis of observational studies. Clin. Lymphoma Myeloma Leuk. 14, 122–130 (2014).
Mitri, J., Castillo, J. & Pittas, A. G. Diabetes and risk of non-Hodgkin’s lymphoma: a metaanalysis of observed studies. Diabetes Care 31, 2391–2397 (2008).
Boyle, T. et al. Physical activity, obesity and survival in diffuse large B cell and follicular lymphoma cases. Br. J. Haematol. 178, 442–447 (2017).
Larsson, S. C. & Wolk, A. Obesity and risk of non-Hodgkin’s lymphoma: a meta-analysis. Int. J. Cancer. 121, 1564–1570 (2007).
Willett, E. V. et al. Non-Hodgkin lymphoma and obesity: a pooled analysis from the InterLymph Consortium. Int. J. Cancer. 122, 2062–2070 (2008).
André, M. P. E. et al. Early positron emission tomography response-adapted treatment in stage I and II Hodgkin lymphoma: final results of the randomized EORTC/LYSA/FIL H10 trial. J. Clin. Oncol. 35, 1786–1794 (2017).
Arai, S. et al. Obesity-associated autoantibody production requires AIM to retain the immunoglobulin M immune complex on follicular dendritic cells. Cell Rep. 3, 1187–1198 (2013).
Winer, D. A. et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat. Med. 17, 610–617 (2011).
Lu, B. et al. Being overweight or obese and risk of developing rheumatoid arthritis among women: a prospective cohort study. Ann. Rheum. Dis. 73, 1914–1922 (2014).
Bennett, B. D. et al. A role for leptin and its cognate receptor in hematopoiesis. Curr. Biol. 6, 1170–1180 (1996).
Lam, Q. L. et al. Leptin signaling maintains B cell homeostasis via induction of Bcl-2 and Cyclin D1. Proc. Natl Acad. Sci. USA 107, 13812–13817 (2010).
Lourenço, E. V. et al. Leptin promotes systemic lupus erythematosus by increasing autoantibody production and inhibiting immune regulation. Proc. Natl Acad. Sci. USA 113, 10637–10642 (2016).
Rongvaux, A. et al. Nicotinamide phosphoribosyl transferase/pre-B cell colony-enhancing factor/visfatin is required for lymphocyte development and cellular resistance to genotoxic stress. J. Immunol. 181, 4685–4695 (2008).
Revollo, J. R. et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 6, 363–375 (2007).
Brentano, F. et al. Pre-B cell colony-enhancing factor/visfatin, a new marker of inflammation in rheumatoid arthritis with proinflammatory and matrix-degrading activities. Arthritis Rheum. 56, 2829–2839 (2007).
Takao, S. et al. Targeting the vulnerability to NAD+ depletion in B cell acute lymphoblastic leukemia. Leukemia 32, 616–625 (2018).
Katerndahl, C. D. S. et al. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat. Immunol. 18, 694–704 (2017).
Schjerven, H. et al. Genetic analysis of Ikaros target genes and tumor suppressor function in BCR-ABL1+ pre-B ALL. J. Exp. Med. 214, 793–814 (2017).
Fretz, J. A. et al. Altered metabolism and lipodystrophy in the early B cell factor 1-deficient mouse. Endocrinology 151, 1611–1621 (2010).
Griffin, M. J. et al. Early B cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J. Biol. Chem. 288, 35925–35939 (2013).
Foley, S. B. et al. Expression of BCR/ABL p210 from a knockin allele enhances bone marrow engraftment without inducing neoplasia. Cell Rep. 17, 51–60 (2013).
Van Nieuwenhove, E. et al. A kindred with mutant IKAROS and autoimmunity. J. Allergy Clin. Immunol. 142, 699–702 (2018).
Wojcik, H. et al. Expression of a non-DNA-binding Ikaros isoform exclusively in B cells leads to autoimmunity but not leukemogenesis. Eur. J. Immunol. 37, 1022–1032 (2007).
Hoshino, A. et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J. Allergy Clin. Immunol. 140, 223–231 (2017).
Papaemmanuil, E. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 41, 1006–1010 (2009).
Trevino, L. R. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 41, 1001–1005 (2009).
Olofsson, L. E. CCAAT/enhancer binding protein alpha (C/EBPalpha) in adipose tissue regulates genes in lipid and glucose metabolism and a genetic variation in C/EBPalpha is associated with serum levels of triglycerides. J. Clin. Endocrinol. Metab. 93, 4880–4886 (2008).
Akasaka, T. et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B cell precursor acute lymphoblastic leukemia (BCPALL). Blood 109, 3451–3461 (2007).
Ryoo, H. et al. Identification of functional nucleotide and haplotype variants in the promoter of the CEBPE gene. J. Hum. Genet. 58, 600–603 (2013).
Okada, Y. et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet. 44, 511–516 (2012).
Yang, W. et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am. J. Hum. Genet. 92, 41–51 (2013).
Lahoud, M. H. et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 11, 1327–1334 (2001).
Cichocki, F. et al. ARID5B regulates metabolic programming in human adaptive NK cells. J. Exp. Med. 215, 2379–2395 (2018).
Olefsky, J. M. Effect of dexamethasone on insulin binding, glucose transport, and glucose oxidation of isolated rat adipocytes. J. Clin. Invest. 56, 1499–1508 (1975).
Marke, R. et al. Tumor suppressor IKZF1 mediates glucocorticoid resistance in B cell precursor acute lymphoblastic leukemia. Leukemia 30, 1599–1603 (2016).
Kaspers, G. J. et al. Different cellular drug resistance profiles in childhood lymphoblastic and non-lymphoblastic leukemia: a preliminary report. Leukemia 8, 1224–1229 (1994).
Geng, H. et al. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell 27, 409–425 (2015).
Feldhahn, N. et al. Mimicry of a constitutively active pre-B cell receptor in acute lymphoblastic leukemia cells. J. Exp. Med. 201, 1837–1852 (2005).
Reth, M. Antigen receptor tail clue. Nature 338, 383–384 (1989).
Khalil, A. M., Cambier, J. C. & Shlomchik, M. J. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science 336, 1178–1181 (2012).
Daëron, M. et al. The same tyrosine-based inhibition motif, in the intracytoplasmic domain of Fc gamma RIIB, regulates negatively BCR-, TCR-, and FcR-dependent cell activation. Immunity 3, 635–646 (1993).
Shojaee, S. Erk negative feedback control enables pre-B cell transformation and represents a therapeutic target in acute lymphoblastic leukemia. Cancer Cell 28, 114–128 (2015).
Hug, E. et al. Inducible expression of hyperactive Syk in B cells activates Blimp-1-dependent terminal differentiation. Oncogene 33, 3730–3741 (2017).
Trageser, D. et al. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J. Exp. Med. 206, 1739–1753 (2009).
Kharabi Masouleh, B. et al. Mechanistic rationale for targeting the unfolded protein response in pre-B acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA 111, E2219–E2228 (2014).
Getahun, A. et al. Continuous inhibitory signaling by both SHP-1 and SHIP-1 pathways is required to maintain unresponsiveness of anergic B cells. J. Exp. Med. 213, 751–769 (2006).
Kersseboom, R. et al. Constitutive activation of Bruton’s tyrosine kinase induces the formation of autoreactive IgM plasma cells. Eur. J. Immunol. 40, 2643–2654 (2010).
Hoyer, B. F. et al. Short-lived plasmablasts and long-lived plasma cells contribute to chronic humoral autoimmunity in NZB/W mice. J. Exp. Med. 199, 1577–1584 (2004).
Negro, R. et al. Overexpression of the autoimmunity-associated phosphatase PTPN22 promotes survival of antigen-stimulated CLL cells by selectively activating AKT. Blood 119, 6278–6287 (2012).
Vang, T. et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet. 37, 1317–1319 (2005).
Hebbring, S. J. et al. Genetic evidence of PTPN22 effects on chronic lymphocytic leukemia. Blood 121, 237–238 (2013).
Schickel, J. N. et al. PTPN22 inhibition resets defective human central B cell tolerance. Sci. Immunol. 1, aaf7153 (2016).
Doughty, C. A. et al. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood 107, 4458–4465 (2006).
Wheeler, M. L. & Defranco, A. L. Prolonged production of reactive oxygen species in response to B cell receptor stimulation promotes B cell activation and proliferation. J. Immunol. 189, 4405–4416 (2012).
Ros, S. & Schulze, A. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 1, 8–12 (2013).
Mouton, V. et al. Heart 6-phosphofructo-2-kinase activation by insulin requires PKB, but not SGK3. Biochem. J. 431, 267–275 (2010).
Bensaad, K. et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 (2006).
Adams, W. C. et al. Anabolism-associated mitochondrial stasis driving lymphocyte differentiation over self-renewal. Cell Rep. 17, 3142–3152 (2016).
Jellusova, J. et al. Gsk3 is a metabolic checkpoint regulator in B cells. Nat. Immunol. 18, 303–312 (2017).
Chen, H. & Chan, D. C. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 26, 39–48 (2017).
Kashatus, J. A. et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 (2015).
Serasinghe, M. N. et al. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol. Cell 57, 521–536 (2015).
Duy, C. et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J. Exp. Med. 207, 1209–1221 (2010).
Nemazee, D. Mechanisms of central tolerance for B xcells. Nat. Rev. Immunol. 17, 281–294 (2017).
Avery, D. T. et al. Germline-activating mutations in PIK3CD compromise B cell development and function. J. Exp. Med. 215, 2073–2095 (2018).
Deau, M. C. et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J. Clin. Invest. 124, 3923–3928 (2014).
Anzelon, A. N., Wu, H. & Rickert, R. C. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat. Immunol. 4, 287–294 (2003).
Browne, C. D. et al. Suppression of phosphatidylinositol 3,4,5-trisphosphate production is a key determinant of B cell anergy. Immunity 31, 749–760 (2009).
Schmitz, R. et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 378, 1396–1407 (2018).
Miletic, A. V. et al. Coordinate suppression of B cell lymphoma by PTEN and SHIP phosphatases. J. Exp. Med. 207, 2407–2420 (2010).
Caro, P. et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 22, 547–560 (2012).
Bordt, E. A. et al. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev. Cell 40, 583–594 (2017).
Nemazee, D. A. & Bürki, K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature 337, 562–566 (1989).
Yurasov, S. et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 201, 703–711 (2005).
Samuels, J. et al. Impaired early B cell tolerance in patients with rheumatoid arthritis. J. Exp. Med. 201, 1659–1667 (2005).
Müschen, M. Rationale for targeting the pre-B cell receptor signaling pathway in acute lymphoblastic leukemia. Blood 125, 3688–3693 (2015).
Young, R. M. & Staudt, L. M. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat. Rev. Drug Discov. 12, 229–243 (2013).
Kojima, H. et al. Differentiation stage-specific requirement in hypoxia-inducible factor-1alpha-regulated glycolytic pathway during murine B cell development in bone marrow. J. Immunol. 184, 154–163 (2010).
Price, M. J. et al. Progressive upregulation of oxidative metabolism facilitates plasmablast differentiation to a T-independent antigen. Cell Rep. 23, 3152–3159 (2018).
Martinez-Martin, N. et al. A switch from canonical to noncanonical autophagy shapes B cell responses. Science 355, 641–647 (2017).
Chen, M. et al. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat. Med. 20, 503–510 (2014).
Clarke, A. J. et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann. Rheum. Dis. 74, 912–920 (2015).
Srinivasan, L. et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell 139, 573–586 (2009).
Tsui, C. et al. Protein kinase C-β dictates B cell fate by regulating mitochondrial remodeling, metabolic reprogramming, and heme biosynthesis. Immunity 48, 1144–1159 (2018).
Patke, A. et al. BAFF controls B cell metabolic fitness through a PKC beta- and Akt-dependent mechanism. J. Exp. Med. 203, 2551–2562 (2006).
Pelanda, R. & Torres, R. M. Central B-cell tolerance: where selection begins. Cold Spring Harb. Perspect. Biol. 4, a007146 (2012).
Sokol, R. J. et al. Human macrophage development: a morphometric study. J. Anat. 151, 27–35 (1987).
Boesen, A. M. Stereologic analysis of the ultrastructure in isolated human T and non-T lymphoid cells. II. Data on blasts in ALL; correlation with immunologic studies and FAB-morphology. Virchows Arch. B 42, 303–314 (1983).
Iwama, Y. & Eguchi, M. Quantitative evaluation of leukemic mitochondria with a computer-controlled image analyser. Virchows Arch. B 51, 375–384 (1986).
Robin, E. D. & Wong, R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell. Physiol. 136, 507–513 (1988).
Siebeneicher, H. et al. Identification and optimization of the first highly selective GLUT1 inhibitor BAY-876. Chem. Med. Chem. 11, 2261–2271 (2016).
Liu, Y. et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol. Cancer Ther. 11, 1672–1682 (2012).
Chan, D. A. et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci. Transl Med. 3, 94ra70 (2011).
Huffman, J. W. et al. 3-(1′,1′-Dimethylbutyl)-1-deoxy-Δ8-THC and related compounds: synthesis of selective ligands for the CB2 receptor. Bioorg. Med. Chem. 7, 2905–2914 (1999).
Kaltenmeier, C. T. et al. Tumor cell-selective inhibitor of mitogen-activated protein kinase phosphatases sensitizes breast cancer cells to lymphokine-activated killer cell activity. J. Pharmacol. Exp. Ther. 361, 39–50 (2017).
Chung, V. et al. Safety, tolerability, and preliminary activity of LB-100, an inhibitor of protein phosphatase 2A, in patients with relapsed solid tumors: An open-label, dose escalation, first-in-human, phase I trial. Clin. Cancer Res. 23, 3277–3284 (2017).
Acknowledgements
The author thanks L. N. Chan, Z. Chen, S. Shojaee, S. Swaminathan, G. Xiao, T. Sadras, G. Deb and other current and former members of his laboratory as well as E. Meffre (New Haven, CT), T. G. Graeber (Los Angeles, CA), A. Weiss and C. A. Lowell (San Francisco, CA), H. Jumaa (Ulm, Germany), A. Melnick (New York, NY) and N. Bottini (La Jolla, CA) for critical discussions and encouragement. M.M. is a Howard Hughes Medical Institute Faculty Scholar (HHMI-55108547) and acknowledges support by the Leukemia and Lymphoma Society (Scholar Award 1479–11), the Wellcome Trust (Senior Investigator Award WT101880) and the US National Cancer Institute (Outstanding Investigator Award R35CA197628).
Reviewer information
Nature Reviews Immunology thanks R. Chiarle, M. Luftig and other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.M. developed the metabolic gatekeeper concept, researched the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Müschen, M. Metabolic gatekeepers to safeguard against autoimmunity and oncogenic B cell transformation. Nat Rev Immunol 19, 337–348 (2019). https://doi.org/10.1038/s41577-019-0154-3
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-019-0154-3
This article is cited by
-
Distinct metabolic requirements regulate B cell activation and germinal center responses
Nature Immunology (2023)
-
PD-1 instructs a tumor-suppressive metabolic program that restricts glycolysis and restrains AP-1 activity in T cell lymphoma
Nature Cancer (2023)
-
Deletion of Glut1 in early postnatal cartilage reprograms chondrocytes toward enhanced glutamine oxidation
Bone Research (2021)