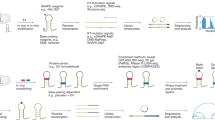
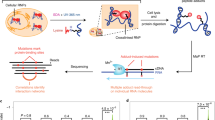
Abstract
RNAs are central to fundamental biological processes in all known organisms. The set of possible intramolecular interactions of RNA nucleotides defines the range of alternative structural conformations of a specific RNA that can coexist, and these structures enable functional catalytic properties of RNAs and/or their productive intermolecular interactions with other RNAs or proteins. However, the immense combinatorial space of potential RNA sequences has precluded predictive mapping between RNA sequence and molecular structure and function. Recent advances in high-throughput approaches in vitro have enabled quantitative thermodynamic and kinetic measurements of RNA–RNA and RNA–protein interactions, across hundreds of thousands of sequence variations. In this Review, we explore these techniques, how they can be used to understand RNA function and how they might form the foundations of an accurate model to predict the structure and function of an RNA directly from its nucleotide sequence. The experimental techniques and modelling frameworks discussed here are also highly relevant for the sampling of sequence–structure–function space of DNAs and proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Tinoco, I. Jr & Bustamante, C. How RNA folds. J. Mol. Biol. 293, 271–281 (1999).
Ganser, L. R., Kelly, M. L., Herschlag, D. & Al-Hashimi, H. M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 20, 474–489 (2019). A comprehensive review that covers how the structural dynamics of RNA control cellular functions.
Al-Hashimi, H. M. & Walter, N. G. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 18, 321–329 (2008).
Winkler, W., Nahvi, A. & Breaker, R. R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Mironov, A. S. et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756 (2002).
Batey, R. T., Gilbert, S. D. & Montange, R. K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432, 411–415 (2004).
Flores, J. K. & Ataide, S. F. Structural changes of RNA in complex with proteins in the SRP. Front. Mol. Biosci. 5, 7 (2018).
Shi, H. et al. Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction. Nat. Commun. 11, 5531 (2020).
Vicens, Q. & Kieft, J. S. Thoughts on how to think (and talk) about RNA structure. Proc. Natl Acad. Sci. USA 119, e2112677119 (2022).
Westhof, E. & Patel, D. J. Nucleic acids. From self-assembly to induced-fit recognition. Curr. Opin. Struct. Biol. 7, 305–309 (1997).
Sussman, J. L., Holbrook, S. R., Warrant, R. W., Church, G. M. & Kim, S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J. Mol. Biol. 123, 607–630 (1978).
Fürtig, B., Richter, C., Wöhnert, J. & Schwalbe, H. NMR spectroscopy of RNA. Chembiochem 4, 936–962 (2003).
Leontis, N. B. & Zirbel, C. L. in RNA 3D Structure Analysis and Prediction (eds Leontis, N. & Westhof, E.) 281–298 (Springer Berlin Heidelberg, 2012).
Holley, R. W. et al. Structure of a ribonucleic acid. Science 147, 1462–1465 (1965).
Peattie, D. A. & Gilbert, W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA 77, 4679–4682 (1980).
Wang, X. D. & Padgett, R. A. Hydroxyl radical ‘footprinting’ of RNA: application to pre-mRNA splicing complexes. Proc. Natl Acad. Sci. USA 86, 7795–7799 (1989).
Latham, J. A. & Cech, T. R. Defining the inside and outside of a catalytic RNA molecule. Science 245, 276–282 (1989).
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M. & Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2017).
Smola, M. J., Rice, G. M., Busan, S., Siegfried, N. A. & Weeks, K. M. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 10, 1643–1669 (2015).
Van Damme, R. et al. Chemical reversible crosslinking enables measurement of RNA 3D distances and alternative conformations in cells. Nat. Commun. 13, 911 (2022).
Hafner, M. et al. CLIP and complementary methods. Nat. Rev. Methods Prim. 1, 1–23 (2021).
Weidmann, C. A., Mustoe, A. M., Jariwala, P. B., Calabrese, J. M. & Weeks, K. M. Analysis of RNA–protein networks with RNP-MaP defines functional hubs on RNA. Nat. Biotechnol. 39, 347–356 (2020).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Spitale, R. C. & Incarnato, D. Probing the dynamic RNA structurome and its functions. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00546-w (2022).
Nutiu, R. et al. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 29, 659–664 (2011). This paper reports the first implementation of a high-throughput biophysical measurement on a sequencing chip, involving binding of the yeast transcription factor GCn4 to a library of DNA sites.
Tome, J. M. et al. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat. Methods 11, 683–688 (2014). This paper reports one of the first implementations of high-throughput biophysical measurements on sequencing chips for RNA, involving the binding of GFP and NELF-E to RNA aptamers.
Buenrostro, J. D. et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat. Biotechnol. 32, 562–568 (2014). This paper reports one of the first implementations of high-throughput biophysical measurements on sequencing chips for RNA, involving binding of the coat protein of MS2 bacteriophage to RNA hairpins.
Layton, C. J., McMahon, P. L. & Greenleaf, W. J. Large-scale, quantitative protein assays on a high-throughput DNA sequencing chip. Mol. Cell 73, 1075–1082.e4 (2019).
Yesselman, J. D. et al. Sequence-dependent RNA helix conformational preferences predictably impact tertiary structure formation. Proc. Natl Acad. Sci. USA 116, 16847–16855 (2019). In this paper, the authors study RNA–RNA binding using tectoRNAs on the RNA array and construct a structure-based model that can predict experimental binding energies.
She, R. et al. Comprehensive and quantitative mapping of RNA–protein interactions across a transcribed eukaryotic genome. Proc. Natl Acad. Sci. USA 114, 3619–3624 (2017).
Li, Z. et al. DNB-based on-chip motif finding: a high-throughput method to profile different types of protein-DNA interactions. Sci. Adv. 6, eabb3350 (2020).
Ozer, A. et al. Quantitative assessment of RNA-protein interactions with high-throughput sequencing–RNA affinity profiling. Nat. Protoc. 10, 1212–1233 (2015).
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014).
Denny, S. K. et al. High-throughput investigation of diverse junction elements in RNA tertiary folding. Cell 174, 377–390.e20 (2018).
Jarmoskaite, I. et al. A quantitative and predictive model for RNA binding by human Pumilio proteins. Mol. Cell 74, 966–981.e18 (2019).
Wu, M. J., Andreasson, J. O. L., Kladwang, W., Greenleaf, W. & Das, R. Automated design of diverse stand-alone riboswitches. ACS Synth. Biol. 8, 1838–1846 (2019).
Becker, W. R. et al. High-throughput analysis reveals rules for target RNA binding and cleavage by AGO2. Mol. Cell 75, 741–755.e11 (2019).
Becker, W. R. et al. Quantitative high-throughput tests of ubiquitous RNA secondary structure prediction algorithms via RNA/protein binding. Preprint at bioRxiv https://doi.org/10.1101/571588 (2019).
Andreasson, J. O. L., Savinov, A., Block, S. M. & Greenleaf, W. J. Comprehensive sequence-to-function mapping of cofactor-dependent RNA catalysis in the glmS ribozyme. Nat. Commun. 11, 1663 (2020).
Bonilla, S. L. et al. High-throughput dissection of the thermodynamic and conformational properties of a ubiquitous class of RNA tertiary contact motifs. Proc. Natl Acad. Sci. USA 118, e2109085118 (2021).
Andreasson, J. O. L. et al. Crowdsourced RNA design discovers diverse, reversible, efficient, self-contained molecular switches. Proc. Natl Acad. Sci. USA 119, e2112979119 (2022).
Jung, C. et al. Massively parallel biophysical analysis of CRISPR-Cas complexes on next generation sequencing chips. Cell 170, 35–47.e13 (2017).
Jones, S. K. Jr et al. Massively parallel kinetic profiling of natural and engineered CRISPR nucleases. Nat. Biotechnol. 39, 84–93 (2021).
Denny, S. K. & Greenleaf, W. J. Linking RNA sequence, structure, and function on massively parallel high-throughput sequencers. Cold Spring Harb. Perspect. Biol. 11, a032300 (2019).
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Cate, J. H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996).
Serganov, A. & Patel, D. J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8, 776–790 (2007).
Glisovic, T., Bachorik, J. L., Yong, J. & Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986 (2008).
Sadée, C. et al. A comprehensive thermodynamic model for RNA binding by the Saccharomyces cerevisiae Pumilio protein PUF4. Nat. Commun. 13, 4522 (2022).
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Boyle, E. A. et al. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl Acad. Sci. USA 114, 5461–5466 (2017).
Ober-Reynolds, B. et al. High-throughput biochemical profiling reveals functional adaptation of a bacterial Argonaute. Mol. Cell 82, 1329–1342.e8 (2022).
Wu, X. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32, 670–676 (2014).
Marklund, E. et al. Sequence specificity in DNA binding is mainly governed by association. Science 375, 442–445 (2022).
Eslami-Mossallam, B. et al. A kinetic model predicts SpCas9 activity, improves off-target classification, and reveals the physical basis of targeting fidelity. Nat. Commun. 13, 1367 (2022). References 56 and 57 (Marklund et al. and Eslami-Mossallam et al.) show how high-throughput data on binding, unbinding and cleavage of DNA by Cas9 can be used to gain microscopic mechanistic insights and build kinetic mechanistic models.
Zadeh, J. N. et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011).
Soukup, G. A. & Breaker, R. R. Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA 96, 3584–3589 (1999).
Suess, B., Fink, B., Berens, C., Stentz, R. & Hillen, W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 32, 1610–1614 (2004).
Förster, T. Zwischenmolekulare energiewanderung und fluoreszenz. Ann. Phys. 437, 55–75 (1948).
Stryer, L. & Haugland, R. P. Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA 58, 719–726 (1967).
Ha, T. Single-molecule fluorescence resonance energy transfer. Methods 25, 78–86 (2001).
Muschielok, A. et al. A nano-positioning system for macromolecular structural analysis. Nat. Methods 5, 965–971 (2008).
Lerner, E. et al. Toward dynamic structural biology: two decades of single-molecule Förster resonance energy transfer. Science 359, eaan1133 (2018).
Chauvier, A. et al. Monitoring RNA dynamics in native transcriptional complexes. Proc. Natl Acad. Sci. USA 118, e2106564118 (2021).
Winz, M.-L., Samanta, A., Benzinger, D. & Jäschke, A. Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry. Nucleic Acids Res. 40, e78 (2012).
Betzig, E. & Chichester, R. J. Single molecules observed by near-field scanning optical microscopy. Science 262, 1422–1425 (1993).
Ha, T. et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl Acad. Sci. USA 93, 6264–6268 (1996).
Zhuang, X. et al. A single-molecule study of RNA catalysis and folding. Science 288, 2048–2051 (2000).
Shema, E. et al. Single-molecule decoding of combinatorially modified nucleosomes. Science 352, 717–721 (2016). This paper shows the first implementation of high-throughput, single-molecule sequencing by synthesis combined with screening of binding, which is used to study nucleosome modifications in a DNA library of the mouse genome.
Severins, I., Joo, C. & van Noort, J. Exploring molecular biology in sequence space: the road to next-generation single-molecule biophysics. Mol. Cell 82, 1788–1805 (2022). This review summarizes the previous applications of high-throughput biophysical measurements on sequencing chips, and discusses in detail how the technology can be extended to carry out single-molecule experiments.
Magde, D., Elson, E. & Webb, W. W. Thermodynamic fluctuations in a reacting system — measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett. 29, 705 (1972).
Yu, L. et al. A comprehensive review of fluorescence correlation spectroscopy. Front. Phys. 9, 644450 (2021).
Zheng, Q. et al. Ultra-stable organic fluorophores for single-molecule research. Chem. Soc. Rev. 43, 1044–1056 (2014).
Marklund, E. et al. DNA surface exploration and operator bypassing during target search. Nature 583, 858–861 (2020).
Wayment-Steele, H. K. et al. RNA secondary structure packages evaluated and improved by high-throughput experiments. Nat. Methods 19, 1234–1242 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Yu, H., Qi, Y. & Ding, Y. Deep learning in RNA structure studies. Front. Mol. Biosci. 9, 869601 (2022).
Singh, J., Hanson, J., Paliwal, K. & Zhou, Y. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nat. Commun. 10, 5407 (2019).
Zhang, H. et al. A new method of RNA secondary structure prediction based on convolutional neural network and dynamic programming. Front. Genet. 10, 467 (2019).
Wang, L. et al. DMfold: a novel method to predict RNA secondary structure with pseudoknots based on deep learning and improved base pair maximization principle. Front. Genet. 10, 143 (2019).
Calonaci, N., Jones, A., Cuturello, F., Sattler, M. & Bussi, G. Machine learning a model for RNA structure prediction. Nar. Genom. Bioinform. 2, lqaa090 (2020).
Sato, K., Akiyama, M. & Sakakibara, Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 12, 941 (2021).
Fu, L. et al. UFold: fast and accurate RNA secondary structure prediction with deep learning. Nucleic Acids Res. 50, e14 (2022).
Townshend, R. J. L. et al. Geometric deep learning of RNA structure. Science 373, 1047–1051 (2021). In this paper, the authors apply deep learning to build a model that can predict the tertiary structure of RNAs after being trained on high-resolution structural data.
Wei, J., Chen, S., Zong, L., Gao, X. & Li, Y. Protein–RNA interaction prediction with deep learning: structure matters. Brief. Bioinform. 23, bbab540 (2021).
Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015).
Lam, J. H. et al. A deep learning framework to predict binding preference of RNA constituents on protein surface. Nat. Commun. 10, 4941 (2019).
Trabelsi, A., Chaabane, M. & Ben-Hur, A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics 35, i269–i277 (2019).
Karniadakis, G. E. et al. Physics-informed machine learning. Nat. Rev. Phys. 3, 422–440 (2021).
Avsec, Ž. et al. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 53, 354–366 (2021).
Arnold, F. H. Design by directed evolution. Acc. Chem. Res. 31, 125–131 (1998).
Arnold, F. H. Combinatorial and computational challenges for biocatalyst design. Nature 409, 253–257 (2001).
Zhao, H., Chockalingam, K. & Chen, Z. Directed evolution of enzymes and pathways for industrial biocatalysis. Curr. Opin. Biotechnol. 13, 104–110 (2002).
Wang, Y., Yu, X. & Zhao, H. Biosystems design by directed evolution. AIChE J. 66, e16716 (2020).
Tan, Z. L. et al. In vivo continuous evolution of metabolic pathways for chemical production. Microb. Cell Fact. 18, 82 (2019).
Wittmann, B. J., Johnston, K. E., Wu, Z. & Arnold, F. H. Advances in machine learning for directed evolution. Curr. Opin. Struct. Biol. 69, 11–18 (2021). This review covers how machine learning has been applied to assist in the navigation of large sequence spaces during directed evolution.
Settles, B. Active learning. Synth. Lect. Artif. Intell. Mach. Learn. 6, 1–114 (2012).
Smith, J. S., Nebgen, B., Lubbers, N., Isayev, O. & Roitberg, A. E. Less is more: sampling chemical space with active learning. J. Chem. Phys. 148, 241733 (2018).
Sverchkov, Y. & Craven, M. A review of active learning approaches to experimental design for uncovering biological networks. PLoS Comput. Biol. 13, e1005466 (2017).
Ennifar, E., Walter, P., Ehresmann, B., Ehresmann, C. & Dumas, P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 8, 1064–1068 (2001).
Okada, K. et al. Solution structure of a GAAG tetraloop in helix 6 of SRP RNA from Pyrococcus furiosus. Nucleosides Nucleotides Nucleic Acids 25, 383–395 (2006).
Kim, N.-K. et al. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J. Mol. Biol. 384, 1249–1261 (2008).
Kuglstatter, A., Oubridge, C. & Nagai, K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat. Struct. Biol. 9, 740–744 (2002).
Stoddard, C. D. et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18, 787–797 (2010).
Collie, G. W., Haider, S. M., Neidle, S. & Parkinson, G. N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 38, 5569–5580 (2010).
Acknowledgements
The authors thank E. Sharma for discussions. This work was supported in part by NIH grants R01GM111990, P50HG007735, R01HG009909, P01GM066275, UM1HG009436 and R01GM121487 to W.J.G. W.J.G. acknowledges support as a Chan Zuckerberg Investigator. E.M. was supported by the Swedish Research Council grant 2020-06459.
Author information
Authors and Affiliations
Contributions
All authors researched, discussed, wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
W.J.G. is a consultant and equity holder for 10x Genomics, Guardant Health, Quantapore and Ultima Genomics, and cofounder of Protillion Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks M. Depken and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Marklund, E., Ke, Y. & Greenleaf, W.J. High-throughput biochemistry in RNA sequence space: predicting structure and function. Nat Rev Genet 24, 401–414 (2023). https://doi.org/10.1038/s41576-022-00567-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-022-00567-5