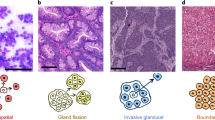
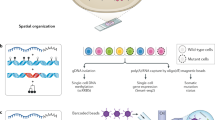
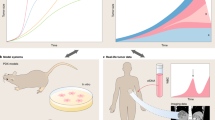
Abstract
The natural history of cancers can be understood through the lens of evolution given that the driving forces of cancer development are mutation and selection of fitter clones. Cancer growth and progression are spatial processes that involve the breakdown of normal tissue organization, invasion and metastasis. For these reasons, spatial patterns are an integral part of histological tumour grading and staging as they measure the progression from normal to malignant disease. Furthermore, tumour cells are part of an ecosystem of tumour cells and their surrounding tumour microenvironment. A range of new spatial genomic, transcriptomic and proteomic technologies offers new avenues for the study of cancer evolution with great molecular and spatial detail. These methods enable precise characterizations of the tumour microenvironment, cellular interactions therein and micro-anatomical structures. In conjunction with spatial genomics, it emerges that tumours and microenvironments co-evolve, which helps explain observable patterns of heterogeneity and offers new routes for therapeutic interventions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cairns, J. Mutation selection and the natural history of cancer. Nature 255, 197–200 (1975). A seminal paper discussing the role of somatic evolution as well as tissue anatomies protective against evolution.
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Martincorena, I. & Campbell, P. J. Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015).
Moore, L. et al. The mutational landscape of human somatic and germline cells. Nature 597, 381–386 (2021). A pan-body atlas of mutations and clonal expansion in normal tissues.
Li, R. et al. A body map of somatic mutagenesis in morphologically normal human tissues. Nature 597, 398–403 (2021).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Greenman, C., Wooster, R., Futreal, P. A., Stratton, M. R. & Easton, D. F. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173, 2187–2198 (2006).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Gonzalez-Perez, A. et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 10, 1081–1082 (2013).
Sondka, Z. et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012). A seminal paper demonstrating that subclonal diversity is common in breast cancer and reconstructing the evolutionary history of copy number alterations.
McGranahan, N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7, 283ra54 (2015).
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2015).
Dentro, S. C. et al. Characterizing genetic intra-tumor heterogeneity across 2658 human cancer genomes. Cell 184, 2239–2254.e39 (2021). A demonstration of universal subclonal divergence across cancer types.
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011). A study pioneering the use of single-cell sequencing for understanding cancer evolution.
Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014).
Casasent, A. K. et al. Multiclonal invasion in breast tumors identified by topographic single. Cell Sequencing. Cell 172, 205–217.e12 (2018).
McPherson, A. et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet. 48, 758–767 (2016).
Laks, E. et al. Clonal decomposition and DNA replication states defined by scaled single-cell genome sequencing. Cell 179, 1207–1221.e22 (2019).
Salehi, S. et al. Clonal fitness inferred from time-series modelling of single-cell cancer genomes. Nature 595, 585–590 (2021).
Williams, M. J., Werner, B., Barnes, C. P., Graham, T. A. & Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 48, 238–244 (2016).
Navin, N. et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 20, 68–80 (2010).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012). One of the first studies using whole-genome sequencing to study patterns of branching and subclonal variegation in kidney cancer.
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Gerlinger, M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014).
Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015).
Morrissy, A. S. et al. Spatial heterogeneity in medulloblastoma. Nat. Genet. 49, 780–788 (2017).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Watkins, T. B. K. et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 587, 126–132 (2020).
Heide, T. et al. The co-evolution of the genome and epigenome in colorectal cancer. Nature 611, 733–743 (2022).
Woodcock, D. J. et al. Prostate cancer evolution from multilineage primary to single lineage metastases with implications for liquid biopsy. Nat. Commun. 11, 5070 (2020).
Grossmann, S. et al. Development, maturation, and maintenance of human prostate inferred from somatic mutations. Cell Stem Cell 28, 1262–1274.e5 (2021).
Zhao, T. et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature 601, 85–91 (2022).
Heide, T. et al. Multiregion human bladder cancer sequencing reveals tumour evolution, bladder cancer phenotypes and implications for targeted therapy. J. Pathol. 248, 230–242 (2019).
Su, F. et al. Spatial intratumor genomic heterogeneity within localized prostate cancer revealed by single-nucleus sequencing. Eur. Urol. 74, 551–559 (2018).
Bao, L. et al. Coexisting genomic aberrations associated with lymph node metastasis in breast cancer. J. Clin. Invest. 128, 2310–2324 (2018).
Househam, J. et al. Phenotypic plasticity and genetic control in colorectal cancer evolution. Nature 611, 744–753 (2022).
Biswas, D. et al. A clonal expression biomarker associates with lung cancer mortality. Nat. Med. 25, 1540–1548 (2019).
Lomakin, A., Svedlund, J., Strell, C., Gataric, M. & Shmatko, A. Spatial genomics maps the structure, character and evolution of cancer clones. Nature 611, 594–602 (2022).
Sottoriva, A. et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Waclaw, B. et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 525, 261–264 (2015).
Gallaher, J. A., Brown, J. S. & Anderson, A. R. A. The impact of proliferation-migration tradeoffs on phenotypic evolution in cancer. Sci. Rep. 9, 2425 (2019).
Lewis, S. M. et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012 (2021).
Lieberman, E., Hauert, C. & Nowak, M. A. Evolutionary dynamics on graphs. Nature 433, 312–316 (2005).
Frean, M., Rainey, P. B. & Traulsen, A. The effect of population structure on the rate of evolution. Proc. Biol. Sci. 280, 20130211 (2013).
Tkadlec, J., Pavlogiannis, A., Chatterjee, K. & Nowak, M. A. Population structure determines the tradeoff between fixation probability and fixation time. Commun. Biol. 2, 138 (2019).
Lopes, J. V., Pacheco, J. M. & Dingli, D. Acquired hematopoietic stem-cell disorders and mammalian size. Blood 110, 4120–4122 (2007).
Lee-Six, H. et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Humphries, A. & Wright, N. A. Colonic crypt organization and tumorigenesis. Nat. Rev. Cancer 8, 415–424 (2008).
Nowak, M. A., Michor, F. & Iwasa, Y. The linear process of somatic evolution. Proc. Natl Acad. Sci. USA 100, 14966–14969 (2003).
Sender, R. & Milo, R. The distribution of cellular turnover in the human body. Nat. Med. 27, 45–48 (2021).
Lee-Six, H. et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574, 532–537 (2019).
Moore, L. et al. The mutational landscape of normal human endometrial epithelium. Nature 580, 640–646 (2020).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). Genovese et al.59 and Jaiswal et al.60 revealed the frequent presence of macroscopic clones with leukaemogenic mutations in the blood of healthy individuals.
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). A pioneering study revealing that normal skin is a patchwork of microscopic mutant clones.
Martincorena, I. et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018).
Lawson, A. R. J. et al. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science 370, 75–82 (2020).
Colom, B. et al. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature 598, 510–514 (2021).
Fowler, J. C. et al. Selection of oncogenic mutant clones in normal human skin varies with body site. Cancer Discov. 11, 340–361 (2021).
Abby, E. et al. Notch1 mutation drives clonal expansion in normal esophageal epithelium but impairs tumor growth. Preprint at bioRxiv https://doi.org/10.1101/2021.06.18.448956 (2021).
Louis, D. N. et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109 (2007).
Elston, C. W. & Ellis, I. O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19, 403–410 (1991).
Epstein, J. I. et al. The 2014 international society of urological pathology (ISUP) consensus conference on gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am. J. Surg. Pathol. 40, 244–252 (2016).
Greenough, R. B. Varying degrees of malignancy in cancer of the breast. J. Cancer Res. 9, 453–463 (1925).
Polyak, K. & Weinberg, R. A. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9, 265–273 (2009).
Morin, P. J. et al. Activation of β-Catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 275, 1787–1790 (1997).
West, J., Schenck, R. O., Gatenbee, C., Robertson-Tessi, M. & Anderson, A. R. A. Normal tissue architecture determines the evolutionary course of cancer. Nat. Commun. 12, 2060 (2021).
Noble, R. et al. Spatial structure governs the mode of tumour evolution. Nat. Ecol. Evol. 6, 207–217 (2022).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Gerstung, M. et al. The evolutionary history of 2658 cancers. Nature 578, 122–128 (2020).
Baslan, T. et al. Ordered and deterministic cancer genome evolution after p53 loss. Nature 608, 795–802 (2022).
Lambert, A. W., Pattabiraman, D. R. & Weinberg, R. A. Emerging biological principles of metastasis. Cell 168, 670–691 (2017).
Fidler, I. J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453–458 (2003).
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288 (2008).
Mamlouk, S. et al. DNA copy number changes define spatial patterns of heterogeneity in colorectal cancer. Nat. Commun. 8, 14093 (2017).
Zhou, H. et al. Multi-region exome sequencing reveals the intratumoral heterogeneity of surgically resected small cell lung cancer. Nat. Commun. 12, 5431 (2021).
Yates, L. R. et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32, 169–184.e7 (2017).
Noorani, A. et al. Genomic evidence supports a clonal diaspora model for metastases of esophageal adenocarcinoma. Nat. Genet. 52, 74–83 (2020).
Karlsson, J. et al. Four evolutionary trajectories underlie genetic intratumoral variation in childhood cancer. Nat. Genet. 50, 944–950 (2018).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Hu, Z., Li, Z., Ma, Z. & Curtis, C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat. Genet. 52, 701–708 (2020).
Brastianos, P. K. et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 5, 1164–1177 (2015).
Brown, D. et al. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat. Commun. 8, 14944 (2017).
De Mattos-Arruda, L. et al. The genomic and immune landscapes of lethal metastatic breast cancer. Cell Rep. 27, 2690–2708.e10 (2019).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Priestley, P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019).
Martínez-Jiménez, F. et al. Pan-cancer whole genome comparison of primary and metastatic solid tumors. Preprint at bioRxiv https://doi.org/10.1101/2022.06.17.496528 (2022).
Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 185, 563–575.e11 (2022). One of the largest genomic analyses of metastasis and organotropism to date.
Reiter, J. G. et al. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 19, 639–650 (2019).
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015).
Somarelli, J. A. The hallmarks of cancer as ecologically driven phenotypes. Front. Ecol. Evol. 9, 661583 (2021).
Darmanis, S. et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 21, 1399–1410 (2017).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, 1391 (2017).
Pombo Antunes, A. R. et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 24, 595–610 (2021).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24 (2017).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Izar, B. et al. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 26, 1271–1279 (2020).
Lambrechts, D. et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018).
Chung, W. et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 8, 15081 (2017).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
Arnol, D., Schapiro, D., Bodenmiller, B., Saez-Rodriguez, J. & Stegle, O. Modeling cell-cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 29, 202–211.e6 (2019).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Raskov, H., Orhan, A., Christensen, J. P. & Gögenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer 124, 359–367 (2021).
Philip, M. & Schietinger, A. CD8+ T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 22, 209–223 (2022).
Borst, J., Ahrends, T., Bąbała, N., Melief, C. J. M. & Kastenmüller, W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 18, 635–647 (2018).
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020).
Sharonov, G. V., Serebrovskaya, E. O., Yuzhakova, D. V., Britanova, O. V. & Chudakov, D. M. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol. 20, 294–307 (2020).
Schumacher, T. N. & Thommen, D. S. Tertiary lymphoid structures in cancer. Science 375, eabf9419 (2022).
Sautès-Fridman, C., Petitprez, F., Calderaro, J. & Fridman, W. H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 19, 307–325 (2019).
Wolf, N. K., Kissiov, D. U. & Raulet, D. H. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat. Rev. Immunol. https://doi.org/10.1038/s41577-022-00732-1 (2022).
Goswami, S., Anandhan, S., Raychaudhuri, D. & Sharma, P. Myeloid cell-targeted therapies for solid tumours. Nat. Rev. Immunol. https://doi.org/10.1038/s41577-022-00737-w (2022).
Murdoch, C., Muthana, M., Coffelt, S. B. & Lewis, C. E. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer 8, 618–631 (2008).
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382 (2019).
Valkenburg, K. C., de Groot, A. E. & Pienta, K. J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 15, 366–381 (2018).
Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16, 582–598 (2016).
Lendahl, U., Muhl, L. & Betsholtz, C. Identification, discrimination and heterogeneity of fibroblasts. Nat. Commun. 13, 3409 (2022).
Sahai, E. et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 20, 174–186 (2020).
Davidson, S. et al. Single-cell RNA sequencing reveals a dynamic stromal niche that supports tumor growth. Cell Rep. 31, 107628 (2020).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 53, 1334–1347 (2021).
Andersson, A. et al. Spatial deconvolution of HER2-positive breast cancer delineates tumor-associated cell type interactions. Nat. Commun. 12, 6012 (2021).
Berglund, E. et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 9, 2419 (2018).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP+fibroblasts and SPP1+macrophages in colorectal cancer. Nat. Commun. 13, 1742 (2022).
Ji, A. L. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 497–514.e22 (2020).
Ravi, V. M. et al. T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat. Commun. 13, 925 (2022).
Wu, R. et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci. Adv. 7, eabg3750 (2021).
Gouin, K. H. III et al. An N-Cadherin 2 expressing epithelial cell subpopulation predicts response to surgery, chemotherapy and immunotherapy in bladder cancer. Nat. Commun. 12, 4906 (2021).
Erickson, A. et al. Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature 608, 360–367 (2022).
Barkley, D. et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet. 54, 1192–1201 (2022).
Danenberg, E. et al. Breast tumor microenvironment structures are associated with genomic features and clinical outcome. Nat. Genet. 54, 660–669 (2022). A detailed spatial proteomic analysis of more than 500 breast cancers revealing recurrent cellular neighbourhoods associated with clinical outcomes.
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Nirmal, A. J. et al. The spatial landscape of progression and immunoediting in primary melanoma at single-cell resolution. Cancer Discov. 12, 1518–1541 (2022). A detailed multiplexed spatial proteomics analysis of melanoma revealing spatially variegated TMEs and cellular interactions.
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e19 (2020).
Wieland, A. et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature 597, 274–278 (2021).
Gaglia, G. et al. Temporal and spatial topography of cell proliferation in cancer. Nat. Cell Biol. 24, 316–326 (2022).
Kather, J. N. et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020).
Fu, Y. et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Loeffler, C. M. L. et al. Artificial intelligence–based detection of FGFR3 mutational status directly from routine histology in bladder cancer: a possible preselection for molecular testing? Eur. Urol. Focus. 8, 472–479 (2022).
Kole, A. J. et al. Overall survival is improved when DCIS accompanies invasive breast cancer. Sci. Rep. 9, 9934 (2019).
Ponz de Leon, M. et al. Evidence for the existence of different types of large bowel tumor: suggestions from the clinical data of a population-based registry. J. Surg. Oncol. 44, 35–43 (1990).
Sawas, T. et al. Identification of prognostic phenotypes of esophageal adenocarcinoma in 2 independent cohorts. Gastroenterology 155, 1720–1728.e4 (2018).
Ross-Innes, C. S. et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat. Genet. 47, 1038–1046 (2015).
Stachler, M. D. et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat. Genet. 47, 1047–1055 (2015).
Thomlinson, R. H. & Gray, L. H. The histological structure of some human lung cancers and the possible implications for radiotherapy. Br. J. Cancer 9, 539–549 (1955).
Hammerl, D. et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat. Commun. 12, 5668 (2021).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387.e19 (2018).
Galon, J. et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 232, 199–209 (2014).
Hegde, P. S. & Chen, D. S. Top 10 challenges in cancer immunotherapy. Immunity 52, 17–35 (2020).
Grünwald, B. T. et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell 184, 5577–5592.e18 (2021).
Tavernari, D. et al. Nongenetic evolution drives lung adenocarcinoma spatial heterogeneity and progression. Cancer Discov. 11, 1490–1507 (2021).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Thorsson, V. et al. The immune landscape of cancer. Immunity 51, 411–412 (2019).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016).
Shukla, S. A. et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 33, 1152–1158 (2015).
Goodman, A. M. et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol. 4, 1237–1244 (2018).
Van den Eynden, J., Jiménez-Sánchez, A., Miller, M. L. & Larsson, E. Lack of detectable neoantigen depletion signals in the untreated cancer genome. Nat. Genet. 51, 1741–1748 (2019).
McGranahan, N. et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017). A seminal study that revealed ample evidence of subclonal immune evasion.
Balachandran, V. P. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017).
Łuksza, M. et al. Neoantigen quality predicts immunoediting in survivors of pancreatic cancer. Nature 606, 389–395 (2022).
Sade-Feldman, M. et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 8, 1136 (2017).
Zaretsky, J. M. et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016).
Dhainaut, M. et al. Spatial CRISPR genomics identifies regulators of the tumor microenvironment. Cell 185, 1223–1239.e20 (2022). A mouse experimental CRISPR model demonstrating direct effects of gene knockouts and TMEs.
Vennin, C. et al. CAF hierarchy driven by pancreatic cancer cell p53-status creates a pro-metastatic and chemoresistant environment via perlecan. Nat. Commun. 10, 3637 (2019).
Tape, C. J. et al. Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell 165, 1818 (2016).
Ali, H. R. et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer 1, 163–175 (2020).
Harris, A. L. Hypoxia — a key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 (2002).
Graeber, T. G. et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379, 88–91 (1996).
Baldominos, P. et al. Quiescent cancer cells resist T cell attack by forming an immunosuppressive niche. Cell 185, 1694–1708.e19 (2022).
Miller, B. E., Miller, F. R. & Heppner, G. H. Therapeutic perturbation of the tumor ecosystem in reconstructed heterogeneous mouse mammary tumors. Cancer Res. 49, 3747–3753 (1989).
Alonso-Curbelo, D. et al. A gene-environment-induced epigenetic program initiates tumorigenesis. Nature 590, 642–648 (2021).
Cleary, A. S., Leonard, T. L., Gestl, S. A. & Gunther, E. J. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 508, 113–117 (2014).
Zhou, H., Neelakantan, D. & Ford, H. L. Clonal cooperativity in heterogenous cancers. Semin. Cell Dev. Biol. 64, 79–89 (2017).
Tammela, T. et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359 (2017).
Williams, J. B. et al. Tumor heterogeneity and clonal cooperation influence the immune selection of IFN-γ-signaling mutant cancer cells. Nat. Commun. 11, 602 (2020).
Diamantopoulou, Z. et al. The metastatic spread of breast cancer accelerates during sleep. Nature 607, 156–162 (2022).
Szczerba, B. M. et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 566, 553–557 (2019).
Adams, D. L. et al. Circulating giant macrophages as a potential biomarker of solid tumors. Proc. Natl Acad. Sci. USA 111, 3514–3519 (2014).
Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 133, 571–573 (1889).
Zeng, Q. et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 573, 526–531 (2019).
Venkataramani, V. et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell 185, 2899–2917.e31 (2022).
Quinn, J. J. et al. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts.Science 371, eabc1944 (2021).
Peinado, H. et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017).
Shiozawa, Y. et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J. Clin. Invest. 121, 1298–1312 (2011).
Kienast, Y. et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122 (2010).
Ghajar, C. M. et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15, 807–817 (2013).
Fane, M. E. et al. Stromal changes in the aged lung induce an emergence from melanoma dormancy. Nature 606, 396–405 (2022).
Reticker-Flynn, N. E. et al. Lymph node colonization induces tumor-immune tolerance to promote distant metastasis. Cell 185, 1924–1942.e23 (2022).
van Maldegem, F. et al. Characterisation of tumour microenvironment remodelling following oncogene inhibition in preclinical studies with imaging mass cytometry. Nat. Commun. 12, 5906 (2021).
Moldoveanu, D. et al. Spatially mapping the immune landscape of melanoma using imaging mass cytometry. Sci. Immunol. 7, eabi5072 (2022).
Erickson, A. et al. Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature 608, 360–367 (2022).
Azimi, F. et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 30, 2678–2683 (2012).
Lee, H. J. et al. Tertiary lymphoid structures: prognostic significance and relationship with tumour-infiltrating lymphocytes in triple-negative breast cancer. J. Clin. Pathol. 69, 422–430 (2016).
Helmink, B. A. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020).
Chen, B. et al. Prognostic value of the common tumour-infiltrating lymphocyte subtypes for patients with non-small cell lung cancer: a meta-analysis. PLoS One 15, e0242173 (2020).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 26, 259–271 (2015).
Lu, S. et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 5, 1195–1204 (2019).
Galon, J. et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006).
Pagès, F. et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 391, 2128–2139 (2018).
Färkkilä, A. et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat. Commun. 11, 1459 (2020).
González-Silva, L., Quevedo, L. & Varela, I. Tumor functional heterogeneity unraveled by scRNA-seq technologies. Trends Cancer Res. 6, 13–19 (2020).
Janiszewska, M. et al. Subclonal cooperation drives metastasis by modulating local and systemic immune microenvironments. Nat. Cell Biol. 21, 879–888 (2019).
Gatenbee, C. D. et al. Immunosuppressive niche engineering at the onset of human colorectal cancer. Nat. Commun. 13, 1798 (2022).
Weinstein, J. N. et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
International Cancer Genome Consortium. International network of cancer genome projects. Nature 464, 993–998 (2010).
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Rozenblatt-Rosen, O. et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Rao, A., Barkley, D., França, G. S. & Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021).
Emmert-Buck, M. R. et al. Laser capture microdissection. Science 274, 998–1001 (1996).
Ellis, P. et al. Reliable detection of somatic mutations in solid tissues by laser-capture microdissection and low-input DNA sequencing. Nat. Protoc. 16, 841–871 (2021).
Nichterwitz, S. et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 7, 12139 (2016).
Cui Zhou, D. et al. Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Nat. Genet. 54, 1390–1405 (2022).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020).
Cho, C.-S. et al. Microscopic examination of spatial transcriptome using Seq-Scope. Cell 184, 3559–3572.e22 (2021).
Deng, Y. et al. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792.e21 (2022).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Lyubimova, A. et al. Single-molecule mRNA detection and counting in mammalian tissue. Nat. Protoc. 8, 1743–1758 (2013).
Chen, F. et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016).
Coskun, A. F. & Cai, L. Dense transcript profiling in single cells by image correlation decoding. Nat. Methods 13, 657–660 (2016).
Codeluppi, S. et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods 15, 932–935 (2018).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Lee, J. H. et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 10, 442–458 (2015).
Qian, X. et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat. Methods 17, 101–106 (2020).
Chen, X., Sun, Y.-C., Church, G. M., Lee, J. H. & Zador, A. M. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 46, e22 (2018).
Alon, S. et al. Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science 371, eaax2656 (2021).
Gyllborg, D. et al. Hybridization-based in situ sequencing (HybISS) for spatially resolved transcriptomics in human and mouse brain tissue. Nucleic Acids Res. 48, e112 (2020).
Su, J.-H., Zheng, P., Kinrot, S. S., Bintu, B. & Zhuang, X. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182, 1641–1659.e26 (2020).
Lu, T., Ang, C. E. & Zhuang, X. Spatially resolved epigenomic profiling of single cells in complex tissues. Cell 185, 4448-4464.e17 (2022).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Ke, R. et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013).
Baker, A.-M. et al. Robust RNA-based in situ mutation detection delineates colorectal cancer subclonal evolution. Nat. Commun. 8, 1998 (2017).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018).
Saka, S. K. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019).
Lin, J.-R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018).
Lin, J.-R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Gerdes, M. J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013).
Keren, L. et al. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 5, eaax5851 (2019).
He, S. et al. High-plex multiomic analysis in FFPE at subcellular level by spatial molecular imaging. Preprint at bioRxiv https://doi.org/10.1101/2021.11.03.467020 (2022).
Bergholtz, H. et al. Best practices for spatial profiling for breast cancer research with the GeoMx® digital spatial profiler. Cancers 13, 4456 (2021).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Mund, A., Brunner, A.-D. & Mann, M. Unbiased spatial proteomics with single-cell resolution in tissues. Mol. Cell 82, 2335–2349 (2022).
Mund, A. et al. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat Biotechnol 40, 1231–1240 (2022).
Palla, G., Fischer, D. S., Regev, A. & Theis, F. J. Spatial components of molecular tissue biology. Nat. Biotechnol. 40, 308–318 (2022).
Lin, J.-R. et al. Multiplexed 3D atlas of state transitions and immune interactions in colorectal cancer. Preprint at bioRxiv https://doi.org/10.1101/2021.03.31.437984 (2021).
Gataric, M. et al. PoSTcode: Probabilistic image-based spatial transcriptomics decoder. Preprint at bioRxiv https://doi.org/10.1101/2021.10.12.464086 (2021).
Dries, R. et al. Advances in spatial transcriptomic data analysis. Genome Res. 31, 1706–1718 (2021).
Perkel, J. M. Starfish enterprise: finding RNA patterns in single cells. Nature 572, 549–551 (2019).
Hickey, J. W. et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods 19, 284–295 (2022).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Nieto, P. et al. A single-cell tumor immune atlas for precision oncology. Genome Res. 31, 1913–1926 (2021).
Park, J. et al. Cell segmentation-free inference of cell types from in situ transcriptomics data. Nat. Commun. 12, 3545 (2021).
Petukhov, V. et al. Cell segmentation in imaging-based spatial transcriptomics. Nat. Biotechnol. 40, 345–354 (2022).
Soldatov, R. et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536 (2019).
Prabhakaran, S. Sparcle: assigning transcripts to cells in multiplexed images. Bioinforma. Adv. 2, vbac048 (2022).
Schapiro, D. et al. MCMICRO: a scalable, modular image-processing pipeline for multiplexed tissue imaging. Nat. Methods 19, 311–315 (2022).
Lohoff, T. et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat. Biotechnol. 40, 74–85 (2022).
Zhang, Y. et al. Reference-based cell type matching of spatial transcriptomics data. Preprint at bioRxiv https://doi.org/10.1101/2022.03.28.486139 (2022).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Kiemen, A. et al. In situ characterization of the 3D microanatomy of the pancreas and pancreatic cancer at single cell resolution. Preprint at bioRxiv https://doi.org/10.1101/2020.12.08.416909 (2020).
Velten, B. et al. Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO. Nat. Methods 19, 179–186 (2022).
Svensson, V., Teichmann, S. A. & Stegle, O. SpatialDE: identification of spatially variable genes. Nat. Methods 15, 343–346 (2018).
Lopez, R. et al. DestVI identifies continuums of cell types in spatial transcriptomics data. Nat. Biotechnol. 40, 1360–1369 (2022).
McCaffrey, E. F. et al. The immunoregulatory landscape of human tuberculosis granulomas. Nat. Immunol. 23, 318–329 (2022).
Dong, K. & Zhang, S. Deciphering spatial domains from spatially resolved transcriptomics with an adaptive graph attention auto-encoder. Nat. Commun. 13, 1739 (2022).
Nagasawa, S. et al. Genomic profiling reveals heterogeneous populations of ductal carcinoma in situ of the breast. Commun. Biol. 4, 438 (2021).
Elyanow, R., Zeira, R., Land, M. & Raphael, B. J. STARCH: copy number and clone inference from spatial transcriptomics data. Phys. Biol. 18, 035001 (2021).
Acknowledgements
We would like to thank S. Dentro, I. Martincorena and T. Grünewald for critical feedback on the manuscript. L.R.Y. is funded by a Wellcome Trust Clinical Research Career Development Fellowship ref: 214584/Z/18/Z.
Author information
Authors and Affiliations
Contributions
Z.S. and A.L. conducted the literature research. Z.S. drew the initial figures based on discussions with all authors. M.G. conceived and supervised the study with L.R.Y. All authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks Alexander Anderson, Alexander Swarbrick, who co-reviewed with Sonny Ramkomuth, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Adjuvant therapies
-
Cancer treatments given after primary therapy (particularly following surgical resection of a tumour) to reduce the risk of relapse.
- Carcinomas in situ
-
Neoplastic expansions of epithelial cells that are confined to the normal tissue structure in which they arose, without invasion of the adjacent stroma.
- Clones
-
A clone is a population of cells that derive from a common ancestor. Cancers are found to be clones deriving from a single cell that expanded during the lifetime of a host. Clonal alterations are mutations that are present in all cells of a cancer because their occurrence preceded the expansion of the tumour.
- Immune-checkpoint blockade
-
A type of anticancer immunotherapy that blocks checkpoint proteins on T cells or their targets on cancer cells and promotes an immune response and killing of the tumour.
- Immune evasion
-
The process by which tumour cells develop several mechanisms that help them to continue to grow and expand by escaping immune control. Immune evasion is thought to be typically preceded by two other phases: elimination, when the immune system recognizes and eliminates tumour cells, and equilibrium, when the pressure from the immune system stalls tumour growth and expansion, and negative selection takes place.
- Invasion
-
The process of penetration and spread of cancer cells into the neighbouring normal tissue. In the case of carcinomas, invasion involves a breach of the basement membrane.
- Metastasis
-
A process of cancer spreading from the primary disease site to lymph nodes or other organs, resulting in the formation of metastatic deposits known as metastases.
- Most recent common ancestor
-
In cancer evolution, the most recent founder cell from which all other tumour cells have directly descended.
- Mutation
-
Changes in the DNA sequence that are inherited across cell generations. Sources of mutation are erroneous replication, biochemical alterations of DNA and failed DNA repair. To date, all cells within the human body are found to accumulate mutations over their lifetime.
- Negative selection
-
The removal of deleterious variants from the population.
- Neoantigen
-
A tumour-specific antigen that is the result of a somatic coding mutation in the corresponding part of a gene. Antigens are protein fragments presented on the cell surface by the human leukocyte antigen (HLA) complex and recognized by different cells of the immune system.
- Positive selection
-
The spread of advantageous alleles within a population.
- Selection
-
In evolution, natural selection denotes the process of survival and reproduction of the fittest organism within a given environment.
- Subclones
-
Further clones emerging within a tumour from one founder clone. Subclonal mutations are limited to a fraction of cancer cells and occur during tumour expansion.
- Tissue architecture
-
The micro-anatomical spatial organization of the tissue. Typical examples are layered epithelial tissues, glands and crypts. Tissue architecture can be combined with other means of tissue organization such as differentiation hierarchies.
- Tumour ecosystems
-
The collective set of heterogeneous cells in the vicinity of a tumour comprising cancer cells and the TME.
- Tumour microenvironment
-
(TME). A combination of non-tumour cells, such as stromal and immune cells, vessels, metabolites, signalling molecules, and other extracellular components among which tumour cells exist.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Seferbekova, Z., Lomakin, A., Yates, L.R. et al. Spatial biology of cancer evolution. Nat Rev Genet 24, 295–313 (2023). https://doi.org/10.1038/s41576-022-00553-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-022-00553-x
This article is cited by
-
Rare subclonal sequencing of breast cancers indicates putative metastatic driver mutations are predominately acquired after dissemination
Genome Medicine (2024)
-
Semi-automated approaches for interrogating spatial heterogeneity of tissue samples
Scientific Reports (2024)
-
Benchmarking spatial clustering methods with spatially resolved transcriptomics data
Nature Methods (2024)
-
SpatialData: an open and universal data framework for spatial omics
Nature Methods (2024)
-
Spatial transcriptomics: recent developments and insights in respiratory research
Military Medical Research (2023)