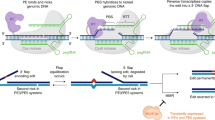
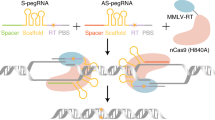
Abstract
Programmable gene-editing tools have transformed the life sciences and have shown potential for the treatment of genetic disease. Among the CRISPR–Cas technologies that can currently make targeted DNA changes in mammalian cells, prime editors offer an unusual combination of versatility, specificity and precision. Prime editors do not require double-strand DNA breaks and can make virtually any substitution, small insertion and small deletion within the DNA of living cells. Prime editing minimally requires a programmable nickase fused to a polymerase enzyme, and an extended guide RNA that both specifies the target site and templates the desired genome edit. In this Review, we summarize prime editing strategies to generate programmed genomic changes, highlight their limitations and recent developments that circumvent some of these bottlenecks, and discuss applications and future directions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 322, 1843–1845 (2008).
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 (2011).
Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816 (2012). This paper reports the use of SpCas9 nuclease and nickases and the development of single-guide RNAs for programmable DNA cutting.
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016).
Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018).
Allen, F. et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 37, 64–72 (2019).
Chen, W. et al. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 47, 7989–8003 (2019).
Iyer, S. et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565 (2019).
Rouet, P., Smih, F. & Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA 91, 6064–6068 (1994).
Rouet, P., Smih, F. & Jasin, M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14, 8096–8106 (1994).
Heyer, W.-D., Ehmsen, K. T. & Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 (2010).
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Cullot, G. et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 10, 1136 (2019).
Alanis-Lobato, G. et al. Frequent loss of heterozygosity in CRISPR-Cas9 edited early human embryos. Proc. Natl Acad. Sci. 118, e2004832117 (2021).
Leibowitz, M. L. et al. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 53, 895–905 (2021).
Tao, J., Wang, Q., Mendez-Dorantes, C., Burns, K. H. & Chiarle, R. Frequency and mechanisms of LINE-1 retrotransposon insertions at CRISPR/Cas9 sites. Nat. Commun. https://doi.org/10.1038/s41467-022-31322-3 (2022).
Ihry, R. J. et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Enache, O. M. et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 52, 662–668 (2020).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). This paper details the development of the first cytosine base editor.
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016).
Gaudelli, N. M. et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). This article describes the engineering and evolution of the first adenine base editor.
Mok, B. Y. et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020).
Cho, S.-I. et al. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell 185, 1764–1776.e1712 (2022).
Yang, L. et al. Engineering and optimising deaminase fusions for genome editing. Nat. Commun. 7, 13330 (2016).
Kurt, I. C. et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 39, 41–46 (2021).
Zhao, D. et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 39, 35–40 (2021).
Chen, L. et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 12, 1384 (2021).
Koblan, L. W. et al. Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 39, 1414–1425 (2021).
Ferrari, S. et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat. Biotechnol. 38, 1298–1308 (2020).
Song, Y. et al. Large-fragment deletions induced by Cas9 cleavage while not in the BEs system. Mol. Ther. Nucleic Acids 21, 523–526 (2020).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Grünewald, J. et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019).
Gehrke, J. M. et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 36, 977–982 (2018).
Kim, Y. B. et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35, 371–376 (2017).
Nishimasu, H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018).
Huang, T. P. et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 37, 626–631 (2019).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020).
Miller, S. M. et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 38, 471–481 (2020).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628 (2020).
Rees, H. A., Wilson, C., Doman, J. L. & Liu, D. R. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 5, eaax5717 (2019).
Grünewald, J. et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 37, 1041–1048 (2019).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). This article describes the original development of prime editing, which includes the PE1, PE2 and PE3 prime editing systems.
Chen, P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e5629 (2021). This article uncovers the inhibitory effect of mismatch repair on prime editing and details the development of the PE4 and PE5 systems, as well as the PEmax architecture.
Nelson, J. W. et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410 (2022). This paper identifies pegRNA degradation as a mechanism that reduces prime editing efficiency and reports the development of epegRNAs with 3ʹ structural motifs that can reduce pegRNA degradation.
Choi, J. et al. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 40, 218–226 (2022). This article describes the PRIME-Del strategy, which uses two pegRNAs to mediate large, targeted deletions.
Anzalone, A. V. et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 40, 731–740 (2022). This article describes the creation of twin prime editing and its use with serine recombinases to enable deletion, replacement, inversion and integration of large sequences.
Jiang, T., Zhang, X.-O., Weng, Z. & Xue, W. Deletion and replacement of long genomic sequences using prime editing. Nat. Biotechnol. 40, 227–234 (2022).
Ioannidi, E. I. et al. Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. Preprint at bioRxiv https://doi.org/10.1101/2021.11.01.466786 (2021).
Ferreira Da Silva, J. et al. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nat. Commun. https://doi.org/10.1038/s41467-022-28442-1 (2022).
Gao, R. et al. Genomic and transcriptomic analyses of prime editing guide RNA–independent off-target effects by prime editors. CRISPR J. 5, 276–293 (2022).
Schene, I. F. et al. Prime editing for functional repair in patient-derived disease models. Nat. Commun. https://doi.org/10.1038/s41467-020-19136-7 (2020).
Geurts, M. H. et al. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Sci. Alliance 4, e202000940 (2021).
Park, S.-J. et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. https://doi.org/10.1186/s13059-021-02389-w (2021).
Liu, Y. et al. Efficient generation of mouse models with the prime editing system. Cell Discov. https://doi.org/10.1038/s41421-020-0165-z (2020).
Gao, P. et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol. https://doi.org/10.1186/s13059-021-02304-3 (2021).
Lin, J. et al. Modeling a cataract disorder in mice with prime editing. Mol. Ther. Nucleic Acids 25, 494–501 (2021).
Jin, S. et al. Genome-wide specificity of prime editors in plants. Nat. Biotechnol. 39, 1292–1299 (2021).
Liu, P. et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. https://doi.org/10.1038/s41467-021-22295-w (2021).
Spencer, J. M. & Zhang, X. Deep mutational scanning of S. pyogenes Cas9 reveals important functional domains. Sci. Rep. https://doi.org/10.1038/s41598-017-17081-y (2017).
Velimirovic, M. et al. Peptide fusion improves prime editing efficiency. Nat. Commun. https://doi.org/10.1038/s41467-022-31270-y (2022).
Song, M. et al. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat. Commun. https://doi.org/10.1038/s41467-021-25928-2 (2021).
Zong, Y. et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 40, 1394–1402 (2022).
Xu, W. et al. A design optimized prime editor with expanded scope and capability in plants. Nat. Plants 8, 45–52 (2022).
Zhang, G. et al. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nat. Commun. https://doi.org/10.1038/s41467-022-29507-x (2022).
Li, X. et al. Enhancing prime editing efficiency by modified pegRNA with RNA G-quadruplexes. J. Mol. Cell Biol. https://doi.org/10.1093/jmcb/mjac022 (2022).
Feng, Y. et al. Enhancing prime editing efficiency and flexibility with tethered and split pegRNAs. Protein Cell https://doi.org/10.1093/procel/pwac014 (2022).
Liu, Y. et al. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 31, 1134–1136 (2021).
Petri, K. et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 40, 189–193 (2022).
Huang, S. et al. Broadening prime editing toolkits using RNA-Pol-II-driven engineered pegRNA. Mol. Ther. 30, 2923–2932 (2022).
Li, X. et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure. Nat. Commun. https://doi.org/10.1038/s41467-022-29339-9 (2022).
Liu, B. et al. A split prime editor with untethered reverse transcriptase and circular RNA template. Nat. Biotechnol. 40, 1388–1393 (2022). This paper describes the split prime editor system and petRNAs, in which the prime editor and pegRNA are each separated into two modular components.
Habib, O., Habib, G., Hwang, G.-H. & Bae, S. Comprehensive analysis of prime editing outcomes in human embryonic stem cells. Nucleic Acids Res. 50, 1187–1197 (2022).
Bothmer, A. et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 8, 13905 (2017).
Liu, N. et al. HDAC inhibitors improve CRISPR/Cas9 mediated prime editing and base editing. Mol. Ther. Nucleic Acids 29, 36–46 (2022).
Kweon, J. et al. Engineered prime editors with PAM flexibility. Mol. Ther. 29, 2001–2007 (2021).
Böck, D. et al. In vivo prime editing of a metabolic liver disease in mice. Sci. Transl. Med. 14, eabl9238 (2022). This article demonstrates the phenotypic rescue of a phenylketonuria mouse model through viral delivery of prime editors.
Oh, Y. et al. Expansion of the prime editing modality with Cas9 from Francisella novicida. Genome Biol. https://doi.org/10.1186/s13059-022-02644-8 (2022).
Zheng, C. et al. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Mol. Ther. 30, 1343–1351 (2022).
Gao, Z. et al. A truncated reverse transcriptase enhances prime editing by split AAV vectors. Mol. Ther. 30, 2942–2951 (2022).
Lin, Q. et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585 (2020).
Doman, J. L., Sousa, A. A., Randolph, P. B., Chen, P. J. & Liu, D. R. Designing and executing prime editing experiments in mammalian cells. Nat. Protoc. https://doi.org/10.1038/s41596-022-00724-4 (2022).
Bhagwat, A. M. et al. Multicrispr: gRNA design for prime editing and parallel targeting of thousands of targets. Life Sci. Alliance 3, e202000757 (2020).
Chow, R. D., Chen, J. S., Shen, J. & Chen, S. A web tool for the design of prime-editing guide RNAs. Nat. Biomed. Eng. 5, 190–194 (2021).
Hwang, G.-H. et al. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 49, W499–W504 (2021).
Li, Y., Chen, J., Tsai, S. Q. & Cheng, Y. Easy-prime: a machine learning–based prime editor design tool. Genome Biol. https://doi.org/10.1186/s13059-021-02458-0 (2021).
Standage-Beier, K., Tekel, S. J., Brafman, D. A. & Wang, X. Prime editing guide RNA design automation using PINE-CONE. ACS Synth. Biol. 10, 422–427 (2021).
Siegner, S. M., Karasu, M. E., Schröder, M. S., Kontarakis, Z. & Corn, J. E. PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinform. https://doi.org/10.1186/s12859-021-04034-6 (2021).
Hsu, J. Y. et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 12, 1034 (2021).
Lin, Q. et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927 (2021). This paper reports the use of dual pegRNAs to increase the efficiency of substitutions, small insertions and small deletions with prime editing.
Kim, H. K. et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 39, 198–206 (2021).
Zhuang, Y. et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat. Chem. Biol. 18, 29–37 (2021).
Wang, J. et al. Efficient targeted insertion of large DNA fragments without DNA donors. Nat. Methods 19, 331–340 (2022).
Tao, R. et al. Bi-PE: bi-directional priming improves CRISPR/Cas9 prime editing in mammalian cells. Nucleic Acids Res. 50, 6423–6434 (2022).
Tao, R. et al. WT-PE: prime editing with nuclease wild-type Cas9 enables versatile large-scale genome editing. Signal Transduct. Target. Ther. https://doi.org/10.1038/s41392-022-00936-w (2022).
Kweon, J. et al. Targeted genomic translocations and inversions generated using a paired prime editing strategy. Mol. Ther. https://doi.org/10.1016/j.ymthe.2022.09.008 (2022).
Adikusuma, F. et al. Optimized nickase- and nuclease-based prime editing in human and mouse cells. Nucleic Acids Res. 49, 10785–10795 (2021).
Peterka, M. et al. Harnessing DSB repair to promote efficient homology-dependent and -independent prime editing. Nat. Commun. https://doi.org/10.1038/s41467-022-28771-1 (2022).
Jang, H. et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 6, 181–194 (2022).
Eggenschwiler, R. et al. A selectable all-in-one CRISPR prime editing piggyBac transposon allows for highly efficient gene editing in human cell lines. Sci. Rep. https://doi.org/10.1038/s41598-021-01689-2 (2021).
Wolff, J. H., Haldrup, J., Thomsen, E. A., Andersen, S. & Mikkelsen, J. G. piggyPrime: high-efficacy prime editing in human cells using piggyBac-based DNA transposition. Front. Genome Ed. https://doi.org/10.3389/fgeed.2021.786893 (2021).
Yuan, Q. & Gao, X. Multiplex base- and prime-editing with drive-and-process CRISPR arrays. Nat. Commun. https://doi.org/10.1038/s41467-022-30514-1 (2022).
Bharucha, N., Ataam, J. A., Gavidia, A. A. & Karakikes, I. Generation of AAVS1 integrated doxycycline-inducible CRISPR-Prime Editor human induced pluripotent stem cell line. Stem Cell Res. 57, 102610 (2021).
Li, H. et al. Highly efficient generation of isogenic pluripotent stem cell models using prime editing. eLife https://doi.org/10.7554/elife.79208 (2022).
Thomas, C. E., Ehrhardt, A. & Kay, M. A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346–358 (2003).
Zhi, S. et al. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 30, 283–294 (2022).
Wang, Q. et al. Broadening the reach and investigating the potential of prime editors through fully viral gene-deleted adenoviral vector delivery. Nucleic Acids Res. 49, 11986–12001 (2021).
Aulicino, F. et al. Highly efficient CRISPR-mediated large DNA docking and multiplexed prime editing using a single baculovirus. Nucleic Acids Res. 50, 7783–7799 (2022).
Sürün, D. et al. Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors. Genes 11, 511 (2020).
Aida, T. et al. Prime editing primarily induces undesired outcomes in mice. Preprint at bioRxiv https://doi.org/10.1101/2020.08.06.239723 (2020).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Landrum, M. J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 (2016).
Chemello, F. et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 7, eabg4910 (2021).
Zhou, M. et al. Targeted-deletion of a tiny sequence via prime editing to restore SMN expression. Int. J. Mol. Sci. 23, 7941 (2022).
Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018).
Li, H. et al. Multiplex precision gene editing by a surrogate prime editor in rice. Mol. Plant. 15, 1077–1080 (2022).
Qian, Y. et al. Efficient and precise generation of Tay–Sachs disease model in rabbit by prime editing system. Cell Discov. https://doi.org/10.1038/s41421-021-00276-z (2021).
Kim, D. E. et al. Prime editor-mediated correction of a pathogenic mutation in purebred dogs. Sci. Rep. https://doi.org/10.1038/s41598-022-17200-4 (2022).
Bosch, J. A., Birchak, G. & Perrimon, N. Precise genome engineering in Drosophila using prime editing. Proc. Natl Acad. Sci. USA 118, e2021996118 (2021).
Doench, J. G. Am I ready for CRISPR? A user’s guide to genetic screens. Nat. Rev. Genet. 19, 67–80 (2018).
Bock, C. et al. High-content CRISPR screening. Nat. Rev. Methods Prim. https://doi.org/10.1038/s43586-021-00093-4 (2022).
Hanna, R. E. et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e1020 (2021).
Kim, Y. et al. High-throughput functional evaluation of human cancer-associated mutations using base editors. Nat. Biotechnol. 40, 874–884 (2022).
Cuella-Martin, R. et al. Functional interrogation of DNA damage response variants with base editing screens. Cell 184, 1081–1097.e1019 (2021).
Erwood, S. et al. Saturation variant interpretation using CRISPR prime editing. Nat. Biotechnol. 40, 885–895 (2022). This paper performs genetic screens and classifies variants of unknown significance using prime editing.
Xu, R., Liu, X., Li, J., Qin, R. & Wei, P. Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice. Nat. Plants 7, 888–892 (2021).
Jiao, Y. et al. Random-PE: an efficient integration of random sequences into mammalian genome by prime editing. Mol. Biomed. 2, 36 (2021).
Oh-hashi, K., Furuta, E., Fujimura, K. & Hirata, Y. Application of a novel HiBiT peptide tag for monitoring ATF4 protein expression in Neuro2a cells. Biochem. Biophys. Rep. 12, 40–45 (2017).
Leonetti, M. D., Sekine, S., Kamiyama, D., Weissman, J. S. & Huang, B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl Acad. Sci. USA 113, E3501–E3508 (2016).
Choi, J. et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing. Nature 608, 98–107 (2022). This article explains the development of the DNA typewriter lineage tracing system, which uses prime editing.
Bortesi, L. & Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52 (2015).
Yin, K., Gao, C. & Qiu, J.-L. Progress and prospects in plant genome editing. Nat. Plants 3, 17107 (2017).
Tang, X. et al. Plant prime editors enable precise gene editing in rice cells. Mol. Plant 13, 667–670 (2020).
Xu, R. et al. Development of plant prime-editing systems for precise genome editing. Plant Commun. 1, 100043 (2020).
Li, J. et al. Development of a highly efficient prime editor 2 system in plants. Genome Biol. https://doi.org/10.1186/s13059-022-02730-x (2022).
Jiang, Y. et al. Optimized prime editing efficiently generates glyphosate-resistant rice plants carrying homozygous TAP-IVS mutation in EPSPS. Mol. Plant. https://doi.org/10.1016/j.molp.2022.09.006 (2022).
Lu, Y. et al. Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol. J. 19, 415–417 (2021).
Perroud, P.-F. et al. Prime editing in the model plant Physcomitrium patens and its potential in the tetraploid potato. Plant Sci. 316, 111162 (2022).
Biswas, S., Bridgeland, A., Irum, S., Thomson, M. J. & Septiningsih, E. M. Optimization of prime editing in rice, peanut, chickpea, and cowpea protoplasts by restoration of GFP activity. Int. J. Mol. Sci. 23, 9809 (2022).
Acknowledgements
D.R.L. acknowledges support from US National Institutes of Health awards U01 AI142756, RM1 HG009490 and R35 GM118062, from the Bill and Melinda Gates Foundation, and from the Howard Hughes Medical Institute. P.J.C. acknowledges support from a US National Science Foundation (NSF) graduate research fellowship. The authors thank A. Anzalone for helpful comments.
Author information
Authors and Affiliations
Contributions
P.J.C. and D.R.L. contributed to all aspects of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have filed patent applications on gene-editing technologies through the Broad Institute of MIT and Harvard. P.J.C. is currently an employee of Prime Medicine. D.R.L. is a consultant and equity owner of Beam Therapeutics, Pairwise Plants, Prime Medicine, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome-editing or genome-engineering technologies.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Chromothripsis
-
A process in which tens to thousands of chromosomal rearrangements occur in a single event.
- Heteroduplex
-
Double-stranded DNA in which the sequences of the strands are not perfectly complementary.
- Mosaicism
-
A condition in which an animal contains multiple cell lineages with different genotypes.
- Nickase Cas9
-
(nCas9). Cas9 that has either its HNH or RuvC nuclease domain catalytically inactivated, resulting in a Cas9 enzyme that can only cut one strand of targeted double-stranded DNA.
- Polygenic diseases
-
Diseases that are mediated by numerous genetic variants that each individually contribute small effects.
- R loop
-
A three-stranded nucleic acid structure that contains a DNA:RNA hybrid and a displaced strand of DNA.
- Single-guide RNA
-
(sgRNA). A single-guide RNA molecule, composed of a CRISPR RNA (crRNA) fused to its corresponding trans-activating CRISPR RNA (tracrRNA) scaffold sequence, that directs the binding and nuclease activity of Cas9 enzymes.
Rights and permissions
About this article
Cite this article
Chen, P.J., Liu, D.R. Prime editing for precise and highly versatile genome manipulation. Nat Rev Genet 24, 161–177 (2023). https://doi.org/10.1038/s41576-022-00541-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-022-00541-1
This article is cited by
-
Improving prime editing with an endogenous small RNA-binding protein
Nature (2024)
-
BacPE: a versatile prime-editing platform in bacteria by inhibiting DNA exonucleases
Nature Communications (2024)
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Template and target-site recognition by human LINE-1 in retrotransposition
Nature (2024)
-
An optimized toolkit for prime editing
Nature Biotechnology (2024)