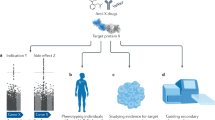
Abstract
Genome-wide association studies (GWAS) have revealed important biological insights into complex diseases, which are broadly expected to lead to the identification of new drug targets and opportunities for treatment. Drug development, however, remains hampered by the time taken and costs expended to achieve regulatory approval, leading many clinicians and researchers to consider alternative paths to more immediate clinical outcomes. In this Review, we explore approaches that leverage common variant genetics to identify opportunities for repurposing existing drugs, also known as drug repositioning. These approaches include the identification of compounds by linking individual loci to genes and pathways that can be pharmacologically modulated, transcriptome-wide association studies, gene-set association, causal inference by Mendelian randomization, and polygenic scoring.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Pushpakom, S. et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41–58 (2019). This review provides a comprehensive overview of the rationale for drug repurposing.
Pammolli, F., Magazzini, L. & Riccaboni, M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 10, 428–438 (2011).
Nosengo, N. Can you teach old drugs new tricks? Nature 534, 314–316 (2016).
Ashburn, T. T. & Thor, K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 (2004).
Verbaanderd, C., Rooman, I., Meheus, L. & Huys, I. On-label or off-label? Overcoming regulatory and financial barriers to bring repurposed medicines to cancer patients. Front. Pharmacol. 10, 1664 (2019).
Watanabe, K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 (2019).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Park, J.-H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
Yang, J., Zeng, J., Goddard, M. E., Wray, N. R. & Visscher, P. M. Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet. 49, 1304–1310 (2017).
Speed, D. et al. Reevaluation of SNP heritability in complex human traits. Nat. Genet. 49, 986–992 (2017).
Pe’er, I., Yelensky, R., Altshuler, D. & Daly, M. J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Fadista, J., Manning, A. K., Florez, J. C. & Groop, L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 24, 1202–1205 (2016).
Paleari, L. et al. Aromatase inhibitors as adjuvant treatment for ER/PgR positive stage I endometrial carcinoma: a retrospective cohort study. Int. J. Mol. Sci. 21, 2227 (2020).
van Weelden, W. J., Massuger, L. F. A. G., ENITEC, Pijnenborg, J. M. A. & Romano, A. Anti-estrogen treatment in endometrial cancer: a systematic review. Front. Oncol. 9, 359 (2019).
O’Mara, T. A. et al. Identification of nine new susceptibility loci for endometrial cancer. Nat. Commun. 9, 3166 (2018).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Stritesky, G. L., Yeh, N. & Kaplan, M. H. IL-23 promotes maintenance but not commitment to the TH17 lineage. J. Immunol. 181, 5948–5955 (2008).
Bunte, K. & Beikler, T. TH17 cells and the IL-23/IL-17 axis in the pathogenesis of periodontitis and immune-mediated inflammatory diseases. Int. J. Mol. Sci. 20, 3394 (2019).
Sandborn, W. J. et al. A randomized trial of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with moderate-to-severe Crohn’s disease. Gastroenterology 135, 1130–1141 (2008).
Sandborn, W. J. et al. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N. Engl. J. Med. 367, 1519–1528 (2012).
Feagan, B. G. et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn’s disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 389, 1699–1709 (2017).
Feagan, B. G. et al. Ustekinumab as induction and maintenance therapy for Crohn’s disease. N. Engl. J. Med. 375, 1946–1960 (2016).
Savage, L. J., Wittmann, M., McGonagle, D. & Helliwell, P. S. Ustekinumab in the treatment of psoriasis and psoriatic arthritis. Rheumatol. Ther. 2, 1–16 (2015).
Banaszczyk, K. Risankizumab in the treatment of psoriasis — literature review. Reumatologia 57, 158–162 (2019).
Singh, S. et al. Selective targeting of the IL23 pathway: generation and characterization of a novel high-affinity humanized anti-IL23A antibody. MAbs 7, 778–791 (2015).
Feagan, B. G. et al. Risankizumab in patients with moderate to severe Crohn’s disease: an open-label extension study. Lancet Gastroenterol. Hepatol. 3, 671–680 (2018).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015). This paper demonstrates the extent to which currently indicated drugs are supported by findings from GWAS.
Beveridge, L. A. et al. Effect of vitamin D supplementation on blood pressure: a systematic review and meta-analysis incorporating individual patient data. JAMA Intern. Med. 175, 745 (2015).
Zhang, D. et al. Effect of vitamin D on blood pressure and hypertension in the general population: an update meta-analysis of cohort studies and randomized controlled trials. Prev. Chronic Dis. 17, E03 (2020).
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Marigorta, U. M. et al. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat. Genet. 49, 1517–1521 (2017).
Morris, J. A. et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 51, 258–266 (2019).
Chandran, T. & Venkatachalam, I. Efficacy and safety of denosumab compared to bisphosphonates in improving bone strength in postmenopausal osteoporosis: a systematic review. Singap. Med. J. 60, 364–378 (2019).
McGovern, D. & Powrie, F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut 56, 1333–1336 (2007).
Hue, S. et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 203, 2473–2483 (2006).
Morris, J. A. et al. Discovery of target genes and pathways of blood trait loci using pooled CRISPR screens and single cell RNA sequencing. Preprint at bioRxiv https://doi.org/10.1101/2021.04.07.438882 (2021).
Nasser, J. et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021).
Schaid, D. J., Chen, W. & Larson, N. B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19, 491–504 (2018). This review provides a comprehensive description of fine-mapping techniques for GWAS signals.
Conrad, D. F. et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 38, 1251–1260 (2006).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
GTEx Consortium et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Reay, W. R. et al. Genetic association and causal inference converge on hyperglycaemia as a modifiable factor to improve lung function. eLife 10, e63115 (2021). This study demonstrates how causal inference can be integrated with the PES approach to support specific repurposing opportunities.
Schizophrenia Working Group of the Psychiatric Genomics Consortium et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 50, 538–548 (2018).
Han, S. et al. Integrating brain methylome with GWAS for psychiatric risk gene discovery. Preprint at bioRxiv https://doi.org/10.1101/440206 (2018).
Zhang, J. et al. Large Bi-ethnic study of plasma proteome leads to comprehensive mapping of cis-pQTL and models for proteome-wide association studies. Preprint at bioRxiv https://doi.org/10.1101/2021.03.15.435533 (2021).
Wingo, A. P. et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat. Genet. 53, 143–146 (2021).
Luningham, J. M. et al. Bayesian genome-wide TWAS method to leverage both cis- and trans-eQTL information through summary statistics. Am. J. Hum. Genet. 107, 714–726 (2020).
Hu, Y. et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 51, 568–576 (2019).
Feng, H. et al. Leveraging expression from multiple tissues using sparse canonical correlation analysis and aggregate tests improves the power of transcriptome-wide association studies. PLoS Genet. 17, e1008973 (2021).
GTEx Consortium et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Gandal, M. J. et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018).
Ratnapriya, R. et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 51, 606–610 (2019).
Wright, G. E. B. et al. Gene expression profiles complement the analysis of genomic modifiers of the clinical onset of Huntington disease. Hum. Mol. Genet. 29, 2788–2802 (2020).
International League Against Epilepsy Consortium on Complex Epilepsies. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun. 9, 5269 (2018).
Gerring, Z. F., Gamazon, E. R., White, A. & Derks, E. M. An integrative network-based analysis reveals gene networks, biological mechanisms, and novel drug targets in Alzheimer’s disease. Preprint at bioRxiv https://doi.org/10.1101/853580 (2019).
Zhang, W. et al. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat. Commun. 10, 3834 (2019).
Zhang, C., Wang, Y., Wang, D., Zhang, J. & Zhang, F. NSAID exposure and risk of Alzheimer’s disease: an updated meta-analysis from cohort studies. Front. Aging Neurosci. 10, 83 (2018).
Heneka, M. T., Reyes-Irisarri, E., Hüll, M. & Kummer, M. P. Impact and therapeutic potential of PPARs in Alzheimer’s disease. Curr. Neuropharmacol. 9, 643–650 (2011).
Musa, A. et al. A review of Connectivity Map and computational approaches in pharmacogenomics. Brief. Bioinform 19, 506–523 (2018).
Wang, Z. et al. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat. Commun. 7, 12846 (2016).
Svoboda, D. L., Saddler, T. & Auerbach, S. S. in Advances in Computational Toxicology Vol. 30 (ed. Hong, H.) 141–157 (Springer, 2019).
Chen, Y.-W. et al. PharmOmics: a species- and tissue-specific drug signature database and online tool for drug repurposing. Preprint at bioRxiv https://doi.org/10.1101/837773 (2019).
Wainberg, M. et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 51, 592–599 (2019). This Perspective comprehensively describes the utility and limitations of the TWAS methodology.
Mancuso, N. et al. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 51, 675–682 (2019).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17 (2017).
eQTLGen, Consortium et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
de Leeuw, C. A., Neale, B. M., Heskes, T. & Posthuma, D. The statistical properties of gene-set analysis. Nat. Rev. Genet. 17, 353–364 (2016). This review summarizes the different approaches and statistical considerations for performing gene-set association.
Liu, J. Z. et al. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 87, 139–145 (2010).
Li, M.-X., Gui, H.-S., Kwan, J. S. H. & Sham, P. C. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 88, 283–293 (2011).
Lamparter, D., Marbach, D., Rueedi, R., Kutalik, Z. & Bergmann, S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput. Biol. 12, e1004714 (2016).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Liu, Y. & Xie, J. Cauchy combination test: a powerful test with analytic P-value calculation under arbitrary dependency structures. J. Am. Stat. Assoc. 115, 393–402 (2020).
de Jong, S., Vidler, L. R., Mokrab, Y., Collier, D. A. & Breen, G. Gene-set analysis based on the pharmacological profiles of drugs to identify repurposing opportunities in schizophrenia. J. Psychopharmacol. 30, 826–830 (2016).
So, H.-C., Chau, C. K.-L., Lau, A., Wong, S.-Y. & Zhao, K. Translating GWAS findings into therapies for depression and anxiety disorders: gene-set analyses reveal enrichment of psychiatric drug classes and implications for drug repositioning. Psychol. Med. 49, 2692–2708 (2019).
Gaspar, H. A. & Breen, G. Drug enrichment and discovery from schizophrenia genome-wide association results: an analysis and visualisation approach. Sci. Rep. 7, 12460 (2017).
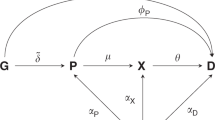
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Slob, E. A. W. & Burgess, S. A comparison of robust Mendelian randomization methods using summary data. Genet. Epidemiol. 44, 313–329 (2020). This study compares different Mendelian randomization methods and their underlying assumptions.
VanderWeele, T. J., Tchetgen Tchetgen, E. J., Cornelis, M. & Kraft, P. Methodological challenges in mendelian randomization. Epidemiology 25, 427–435 (2014).
Burgess, S., Bowden, J., Fall, T., Ingelsson, E. & Thompson, S. G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology 28, 30–42 (2017).
Burgess, S. et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 30, 543–552 (2015).
Burgess, S. et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 4, 186 (2019).
McGowan, L. M., Davey Smith, G., Gaunt, T. R. & Richardson, T. G. Integrating Mendelian randomization and multiple-trait colocalization to uncover cell-specific inflammatory drivers of autoimmune and atopic disease. Hum. Mol. Genet. 28, 3293–3300 (2019).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Hormozdiari, F. et al. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99, 1245–1260 (2016).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Suhre, K., McCarthy, M. I. & Schwenk, J. M. Genetics meets proteomics: perspectives for large population-based studies. Nat. Rev. Genet. https://doi.org/10.1038/s41576-020-0268-2 (2020).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Zheng, J. et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. https://doi.org/10.1038/s41588-020-0682-6 (2020). This study demonstrates how pQTLs could be utilized through Mendelian randomization to inform drug repurposing.
Schmidt, A. F. et al. Genetic drug target validation using Mendelian randomisation. Nat. Commun. 11, 3255 (2020).
Folkersen, L. et al. Genomic evaluation of circulating proteins for drug target characterisation and precision medicine. Preprint at bioRxiv https://doi.org/10.1101/2020.04.03.023804 (2020).
Suhre, K. et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 8, 14357 (2017).
Mokry, L. E. et al. Vitamin D and risk of multiple sclerosis: a mendelian randomization study. PLoS Med. 12, e1001866 (2015).
Lotta, L. A. et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med. 13, e1002179 (2016).
Aikens, R. C. et al. Systolic blood pressure and risk of type 2 diabetes: a Mendelian randomization study. Diabetes 66, 543–550 (2017).
Yin, P. et al. Serum calcium and risk of migraine: a Mendelian randomization study. Hum. Mol. Genet. 26, 820–828 (2016).
Adams, D. M., Reay, W. R., Geaghan, M. P. & Cairns, M. J. Investigation of glycaemic traits in psychiatric disorders using Mendelian randomisation revealed a causal relationship with anorexia nervosa. Neuropsychopharmacology 46, 1093–1102 (2020).
Koellinger, P. D. & de Vlaming, R. Mendelian randomization: the challenge of unobserved environmental confounds. Int. J. Epidemiol. 48, 665–671 (2019).
Gkatzionis, A. & Burgess, S. Contextualizing selection bias in Mendelian randomization: how bad is it likely to be? Int. J. Epidemiol. 48, 691–701 (2019).
O’Connor, L. J. & Price, A. L. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat. Genet. 50, 1728–1734 (2018). This study reveals that genetic correlation can bias Mendelian randomization and provides a novel causal inference method, which explicitly models genetic correlation, to overcome this.
International Schizophrenia Consortium et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Wray, N. R., Goddard, M. E. & Visscher, P. M. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 17, 1520–1528 (2007).
Xue, A. et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 9, 2941 (2018).
Arnedo, J. et al. PGMRA: a web server for (phenotype x genotype) many-to-many relation analysis in GWAS. Nucleic Acids Res. 41, W142–W149 (2013).
Hari Dass, S. A. et al. A biologically-informed polygenic score identifies endophenotypes and clinical conditions associated with the insulin receptor function on specific brain regions. EBioMedicine 42, 188–202 (2019).
Reay, W. R., Atkins, J. R., Carr, V. J., Green, M. J. & Cairns, M. J. Pharmacological enrichment of polygenic risk for precision medicine in complex disorders. Sci. Rep. 10, 879 (2020). This study describes the rationale for the PES approach.
Ghoussaini, M. et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 49, D1311–D1320 (2021).
Fang, H. et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 51, 1082–1091 (2019). This study demonstrates how individual GWAS loci can be integrated with systems biology to repurpose drugs for immunological disorders.
Sakaue, S. & Okada, Y. GREP: Genome for REPositioning drugs. Bioinformatics 35, 3821–3823 (2019).
Gaspar, H. A., Hübel, C. & Breen, G. Drug Targetor: a web interface to investigate the human druggome for over 500 phenotypes. Bioinformatics 35, 2515–2517 (2019).
Konuma, T., Ogawa, K. & Okada, Y. Integration of genetically regulated gene expression and pharmacological library provides therapeutic drug candidates. Hum. Mol. Genet. 30, 294–304 (2021).
Emon, M. A., Domingo-Fernández, D., Hoyt, C. T. & Hofmann-Apitius, M. PS4DR: a multimodal workflow for identification and prioritization of drugs based on pathway signatures. BMC Bioinforma. 21, 231 (2020).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Freshour, S. et al. Integration of the Drug–Gene Interaction Database (DGIdb) with open crowdsource efforts. Preprint at biorxiv https://doi.org/10.1101/2020.09.18.301721 (2020).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Wu, M. C. et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89, 82–93 (2011).
Cordell, H. J. Detecting gene–gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404 (2009).
McAllister, K. et al. Current challenges and new opportunities for gene–environment interaction studies of complex diseases. Am. J. Epidemiol. 186, 753–761 (2017).
Hopkins, A. L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 4, 682–690 (2008).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Michailidou, K. et al. Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94 (2017).
Estrada, K. et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44, 491–501 (2012).
Eyre, S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012).
Evangelou, E. et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425 (2018).
Nair, R. P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 41, 199–204 (2009).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Nielsen, J. B. et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 50, 1234–1239 (2018).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Hartwig, F. P., Davey Smith, G. & Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998 (2017).
Burgess, S., Foley, C. N., Allara, E., Staley, J. R. & Howson, J. M. M. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat. Commun. 11, 376 (2020).
Pierce, B. L., Ahsan, H. & VanderWeele, T. J. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 40, 740–752 (2011).
Bowden, J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR–Egger regression: the role of the I2 statistic. Int. J. Epidemiol. 45, 1961–1974 (2016).
Burgess, S., Zuber, V., Gkatzionis, A. & Foley, C. N. Modal-based estimation via heterogeneity-penalized weighting: model averaging for consistent and efficient estimation in Mendelian randomization when a plurality of candidate instruments are valid. Int. J. Epidemiol. 47, 1242–1254 (2018).
Verbanck, M., Chen, C.-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Mavaddat, N. et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 104, 21–34 (2019).
Acknowledgements
M.J.C. is supported by a National Health and Medical Research Council (NHMRC) Senior Research Fellowship (1121474) and a University of Newcastle Faculty of Health and Medicine Gladys M Brawn Senior Fellowship. W.R.R. is supported by an Australian government research training programme stipend.
Author information
Authors and Affiliations
Contributions
W.R.R. researched the literature. The authors contributed equally to all other aspects of the article.
Corresponding author
Ethics declarations
Competing interests
W.R.R. and M.J.C. have filed a patent related to the use of the pharmagenic enrichment score (PES) framework in complex disorders (WIPO Patent Application WO/2020/237314).
Additional information
Peer review information
Nature Reviews Genetics thanks S. Burgess and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Polygenic
-
A term that denotes the contribution of many genes to the genetic component of a trait.
- Genome-wide association studies
-
(GWAS). Studies using a design that tests the association (relationship) between sequence nucleotide alterations (genetic variants) throughout the genome with a trait of interest, such as a disease phenotype.
- Pleiotropy
-
A term to denote the influence of a gene or genetic variant on multiple different biological traits.
- Imputation
-
Using genetic variants to predict (impute) a particular variable.
- Heritability
-
The proportion of variance in a phenotype in a population that is explained by genetic variation.
- Gene-set association
-
A technique that examines whether a set of genes is associated with a trait by combining the association of individual genetic variants within the set.
- Single-nucleotide polymorphism
-
(SNP). A single-nucleotide alteration in the genomic sequence at any given position (locus).
- Quantitative trait loci
-
Genetic variants or intervals that are linked to or associated with a quantitative trait (measurable continuous phenotype); for example, expression quantitative trait loci (eQTLs) are variants associated with mRNA expression for a given gene.
- Linkage disequilibrium
-
Genetic variants that are inherited together at a higher rate than by chance alone are said to exhibit linkage disequilibrium.
- Fine-mapping
-
Investigating which genetic variant or variants within a region of the genome significantly associated with a trait (genome-wide association study (GWAS) locus) causally influence the trait in question, rather than merely being inherited with the causal variant(s) through linkage disequilibrium.
- Transcriptome-wide association study
-
(TWAS). A technique that tests the association between the predicted expression of a gene based on genetic variants from expression quantitative trait loci (eQTLs) analysis in an independent cohort and a trait of interest.
- Mendelian randomization
-
Randomization using single-nucleotide polymorphisms (SNPs) as instrumental variables (IVs) (proxies of a trait, termed the exposure) to test the causal effect of that trait on another (termed the outcome).
- Biological pathways
-
Genes whose products exert biologically related functions or interact together.
- Instrumental variables
-
(IVs). Independent variables that are used to evaluate whether an exposure causes an outcome or is simply correlated with it.
- Polygenic score
-
A sum of the effect sizes of genetic variants throughout the genome for a particular trait.
- Pharmagenic enrichment score
-
(PES). A polygenic score that is constructed from variants specifically within a biological pathway that is targeted by an approved drug, rather than genome-wide like a traditional polygenic score.
Rights and permissions
About this article
Cite this article
Reay, W.R., Cairns, M.J. Advancing the use of genome-wide association studies for drug repurposing. Nat Rev Genet 22, 658–671 (2021). https://doi.org/10.1038/s41576-021-00387-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-021-00387-z
This article is cited by
-
Genetic influences on circulating retinol and its relationship to human health
Nature Communications (2024)
-
Unveiling potential drug targets for hyperparathyroidism through genetic insights via Mendelian randomization and colocalization analyses
Scientific Reports (2024)
-
Potential drug targets for gastroesophageal reflux disease and Barrett’s esophagus identified through Mendelian randomization analysis
Journal of Human Genetics (2024)
-
Refining the impact of genetic evidence on clinical success
Nature (2024)
-
Priority index for critical Covid-19 identifies clinically actionable targets and drugs
Communications Biology (2024)