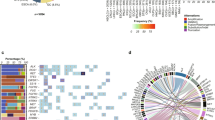
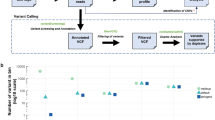
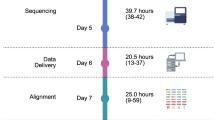
Abstract
Technological innovation and rapid reduction in sequencing costs have enabled the genomic profiling of hundreds of cancer-associated genes as a component of routine cancer care. Tumour genomic profiling can refine cancer subtype classification, identify which patients are most likely to benefit from systemic therapies and screen for germline variants that influence heritable cancer risk. Here, we discuss ongoing efforts to enhance the clinical utility of tumour genomic profiling by integrating tumour and germline analyses, characterizing allelic context and identifying mutational signatures that influence therapy response. We also discuss the potential clinical utility of more comprehensive whole-genome and whole-transcriptome sequencing and ultra-sensitive cell-free DNA profiling platforms, which allow for minimally invasive, serial analyses of tumour-derived DNA in blood.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sobin, L. H. The international histological classification of tumours. Bull. World Health Organ. 59, 813–819 (1981).
Hoadley, K. A. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014).
Bouwman, P. & Jonkers, J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer 12, 587–598 (2012).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Hyman, D. M. et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 373, 726–736 (2015).
Drilon, A. et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med. 378, 731–739 (2018).
Hyman, D. M. et al. AKT inhibition in solid tumors with AKT1 mutations. J. Clin. Oncol. 35, 2251–2259 (2017).
Hungerford, D. A. & Nowell, P. C. A minute chromosome in human chronic granulocytic leukemia. Science 132, 1497–1499 (1960).
de Klein, A. et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature 300, 765–767 (1982).
Kantarjian, H. et al. Improved survival in chronic myeloid leukemia since the introduction of imatinib therapy: a single-institution historical experience. Blood 119, 1981–1987 (2012).
Prasad, V. Perspective: the precision-oncology illusion. Nature 537, S63 (2016).
Hyman, D. M., Taylor, B. S. & Baselga, J. Implementing genome-driven oncology. Cell 168, 584–599 (2017).
Pan, Y. et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer 84, 121–126 (2014).
Mansfield, E. A. FDA perspective on companion diagnostics: an evolving paradigm. Clin. Cancer Res. 20, 1453–1457 (2014).
Kris, M. G. et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 311, 1998–2006 (2014).
Jordan, E. J. et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 7, 596–609 (2017).
Su, Z. et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J. Mol. Diagn. 13, 74–84 (2011).
MacConaill, L. E. et al. Prospective enterprise-level molecular genotyping of a cohort of cancer patients. J. Mol. Diagn. 16, 660–672 (2014).
Li, T., Kung, H.-J., Mack, P. C. & Gandara, D. R. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J. Clin. Oncol. 31, 1039–1049 (2013).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Sholl, L. M. et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 1, e87062 (2016).
Meric-Bernstam, F. et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J. Clin. Oncol. 33, 2753–2762 (2015).
Stockley, T. L. et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 8, 109 (2016).
Odegaard, J. I. et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin. Cancer Res. 24, 3539–3549 (2018).
Brannon, A. R. et al. Enhanced specificity of high sensitivity somatic variant profiling in cell-free DNA via paired normal sequencing: design, validation, and clinical experience of the MSK-ACCESS liquid biopsy assay. Preprint at bioRxiv https://doi.org/10.21203/rs.3.rs-120695/v1 (2020).
Tao, J. J., Schram, A. M. & Hyman, D. M. Basket studies: redefining clinical trials in the era of genome-driven oncology. Annu. Rev. Med. 69, 319–331 (2018).
Redig, A. J. & Jänne, P. A. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J. Clin. Oncol. 33, 975–977 (2015).
Woodcock, J. & LaVange, L. M. Master protocols to study multiple therapies, multiple diseases, or both. N. Engl. J. Med. 377, 62–70 (2017).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Drilon, A. E. et al. A phase II basket study of the oral TRK inhibitor LOXO-101 in adult subjects with NTRK fusion-positive tumors. J. Clin. Oncol. 34, TPS2599–TPS2599 (2016).
Berger, M. F. & Mardis, E. R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 15, 353–365 (2018).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Mateo, J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015).
Van Allen, E. M. et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 4, 1140–1153 (2014).
Li, Q. et al. ERCC2 helicase domain mutations confer nucleotide excision repair deficiency and drive cisplatin sensitivity in muscle-invasive bladder cancer. Clin. Cancer Res. 25, 977–988 (2019).
Kelderman, S., Schumacher, T. N. & Kvistborg, P. Mismatch repair-deficient cancers are targets for anti-PD-1 therapy. Cancer Cell 28, 11–13 (2015).
Chang, M. T. et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016).
Chang, M. T. et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 8, 174–183 (2018).
Hanrahan, A. J. et al. Leveraging systematic functional analysis to benchmark an in silico framework distinguishes driver from passenger MEK mutants in cancer. Cancer Res. 80, 4233–301 (2020).
Hess, J. M. et al. Passenger hotspot mutations in cancer. Cancer Cell 36, 288–301.e14 (2019).
Buisson, R. et al. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science 364, eaaw2872 (2019).
Holbrook, J. A., Neu-Yilik, G., Hentze, M. W. & Kulozik, A. E. Nonsense-mediated decay approaches the clinic. Nat. Genet. 36, 801–808 (2004).
Cheung, L. W. T. et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 1, 170–185 (2011).
Cheung, L. W. T. et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell 26, 479–494 (2014).
Yao, Z. et al. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28, 370–383 (2015).
Zabransky, D. J. et al. HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc. Natl Acad. Sci. USA 112, E6205–E6214 (2015).
Poulikakos, P. I., Zhang, C., Bollag, G., Shokat, K. M. & Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010).
Kopetz, S. et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med. 381, 1632–1643 (2019).
Corcoran, R. B. et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 8, 428–443 (2018).
Gray, S. W., Hicks-Courant, K., Cronin, A., Rollins, B. J. & Weeks, J. C. Physicians’ attitudes about multiplex tumor genomic testing. J. Clin. Oncol. 32, 1317–1323 (2014).
Schram, A. M. et al. Oncologist use and perception of large panel next-generation tumor sequencing. Ann. Oncol. 28, 2298–2304 (2017).
Wu, J.-Y. et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin. Cancer Res. 14, 4877–4882 (2008).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00011 (2017).
Griffith, M. et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017).
Patterson, S. E. et al. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum. Genomics 10, 4 (2016).
Huang, L. et al. The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J. Am. Med. Inform. Assoc. 24, 513–519 (2017).
Tamborero, D. et al. Cancer genome interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 10, 25 (2018).
Dumbrava, E. I. & Meric-Bernstam, F. Personalized cancer therapy-leveraging a knowledge base for clinical decision-making. Cold Spring Harb. Mol. Case Stud. 4, a001578 (2018).
Iyer, G. et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 338, 221 (2012).
Ross, J. S. et al. Comprehensive genomic profiling of carcinoma of unknown primary site: new routes to targeted therapies. JAMA Oncol. 1, 40–49 (2015).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Li, M. M. et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 19, 4–23 (2017).
Mateo, J. et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets (ESCAT). Ann. Oncol. 29, 1895–1902 (2018).
AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831 (2017).
Wagner, A. H. et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 52, 448–457 (2020).
Ritter, D. I. et al. Somatic cancer variant curation and harmonization through consensus minimum variant level data. Genome Med. 8, 117 (2016).
Center for Devices & Radiological Health. FDA recognition of public human genetic variant databases. FDA https://www.fda.gov/medical-devices/precision-medicine/fda-recognition-public-human-genetic-variant-databases (2019).
Stein, E. M. et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731 (2017).
Pollyea, D. A. et al. Enasidenib, an inhibitor of mutant IDH2 proteins, induces durable remissions in older patients with newly diagnosed acute myeloid leukemia. Leukemia 33, 2575–2584 (2019).
DiNardo, C. D. et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 378, 2386–2398 (2018).
André, F. et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019).
Loriot, Y. et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 381, 338–348 (2019).
Abou-Alfa, G. K. et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 21, 671–684 (2020).
Drilon, A. et al. PL02.08 registrational results of LIBRETTO-001: a phase 1/2 trial of LOXO-292 in patients with RET fusion-positive lung cancers. J. Thorac. Oncol. 14, S6–S7 (2019).
Wirth, L. et al. LBA93 - registrational results of LOXO-292 in patients with RET-altered thyroid cancers. Ann. Oncol. 30, v933 (2019).
Litton, J. K. et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 379, 753–763 (2018).
Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019).
Mateo, J. et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 21, 162–174 (2020).
Planchard, D. et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 18, 1307–1316 (2017).
Diamond, E. L. et al. Vemurafenib for BRAF V600-mutant erdheim-chester disease and langerhans cell histiocytosis: analysis of data from the histology-independent, phase 2, open-label VE-BASKET study. JAMA Oncol. 4, 384–388 (2018).
Subbiah, V. et al. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer. J. Clin. Oncol. 36, 7–13 (2018).
Goodman, A. M. et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 16, 2598–2608 (2017).
Amado, R. G. et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 1626–1634 (2008).
De Roock, W. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 11, 753–762 (2010).
Chung, V. et al. Effect of selumetinib and MK-2206 vs oxaliplatin and fluorouracil in patients with metastatic pancreatic cancer after prior therapy: SWOG S1115 study randomized clinical trial. JAMA Oncol. 3, 516–522 (2017).
Jänne, P. A. et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: the SELECT-1 randomized clinical trial. JAMA 317, 1844–1853 (2017).
Canon, J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019).
Tonin, P. et al. BRCA1 mutations in Ashkenazi Jewish women. Am. J. Hum. Genet. 57, 189 (1995).
Abeliovich, D. et al. The founder mutations 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 appear in 60% of ovarian cancer and 30% of early-onset breast cancer patients among Ashkenazi women. Am. J. Hum. Genet. 60, 505–514 (1997).
Newman, B., Austin, M. A., Lee, M. & King, M. C. Inheritance of human breast cancer: evidence for autosomal dominant transmission in high-risk families. Proc. Natl Acad. Sci. USA 85, 3044–3048 (1988).
Hall, J. M. et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684–1689 (1990).
King, M.-C. ‘The race’ to clone BRCA1. Science 343, 1462–1465 (2014).
Miki, Y. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994).
Mandelker, D. et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA 318, 825–835 (2017).
Garofalo, A. et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 8, 79 (2016).
Jones, S. et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci. Transl. Med. 7, 283ra53 (2015).
Zhang, J. et al. Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med. 373, 2336–2346 (2015).
Schrader, K. A. et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2, 104–111 (2016).
Domchek, S. M. Germline genetic testing for breast cancer: Which patients? What genes? Genet. Med. 22, 698–700 (2020).
Konstantinopoulos, P. A. et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J. Clin. Oncol. 38, 1222–1245 (2020).
McLeod, H. L. Cancer pharmacogenomics: early promise, but concerted effort needed. Science 339, 1563–1566 (2013).
Wang, L., McLeod, H. L. & Weinshilboum, R. M. Genomics and drug response. N. Engl. J. Med. 364, 1144–1153 (2011).
Irvin, W. J. Jr. et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: a multicenter study. J. Clin. Oncol. 29, 3232–3239 (2011).
Hertz, D. L. et al. CYP2C8* 3 predicts benefit/risk profile in breast cancer patients receiving neoadjuvant paclitaxel. Breast Cancer Res. Treat. 134, 401–410 (2012).
Pullarkat, S. T. et al. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 1, 65–70 (2001).
Coombs, C. C. et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382.e4 (2017).
Ptashkin, R. N. et al. Prevalence of clonal hematopoiesis mutations in tumor-only clinical genomic profiling of solid tumors. JAMA Oncol. 4, 1589–1593 (2018).
Razavi, P. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019).
Carpenter, E. L. & Mossé, Y. P. Targeting ALK in neuroblastoma — preclinical and clinical advancements. Nat. Rev. Clin. Oncol. 9, 391–399 (2012).
Wirth, L. J. et al. 1922P Exploratory patient-reported outcomes among patients with RET-mutant medullary thyroid cancer in LIBRETTO-001: a phase I/II trial of selpercatinib (LOXO-292). Ann. Oncol. 31 (Suppl. 4), S1089 (2020).
Wells, S. A. Jr. et al. Vandetanib in patients with locally advanced or metastatic medullary thyroid cancer: a randomized, double-blind phase III trial. J. Clin. Oncol. 30, 134–141 (2012).
Yurgelun, M. B. & Hampel, H. Recent advances in Lynch syndrome: diagnosis, treatment, and cancer prevention. Am. Soc. Clin. Oncol. Educ. Book 38, 101–109 (2018).
Rodrigues, M. et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat. Commun. 9, 1866 (2018).
Johansson, P. A. et al. Correction to: Prolonged stable disease in a uveal melanoma patient with germline MBD4 nonsense mutation treated with pembrolizumab and ipilimumab. Immunogenetics 71, 511 (2019).
Schneid, T. in Discrimination Law Issues for the Safety Professional (ed. Schneid, T.) 161–194 (CRC Press, 2011).
National Human Genome Research Institute. Genetic Information Nondiscrimination Act (GINA) of 2008. NIH https://www.genome.gov/24519851/genetic-information-nondiscrimination-act-of-2008 (2008).
Gniady, J. A. Regulating direct-to-consumer genetic testing: protecting the consumer without quashing a medical revolution. Fordham Law Rev. 76, 2429–2475 (2008).
Ferreira-Gonzalez, A. et al. US system of oversight for genetic testing: a report from the Secretary’s Advisory Committee on Genetics, Health and Society. Per. Med. 5, 521–528 (2008).
Lolkema, M. P. et al. Ethical, legal, and counseling challenges surrounding the return of genetic results in oncology. J. Clin. Oncol. 31, 1842–1848 (2013).
Li, M. M. et al. Points to consider for reporting of germline variation in patients undergoing tumor testing: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 22, 1142–1148 (2020).
Kurian, A. W. et al. Genetic testing and counseling among patients with newly diagnosed breast cancer. JAMA 317, 531–534 (2017).
McNamara, D. Shortage of genetic counselors in face of growing need. Medscape https://www.medscape.com/viewarticle/877135 (2017).
Eisen, A. et al. Genetic assessment wait time indicators in the high risk ontario breast screening program. Mol. Genet. Genomic Med. 6, 213–223 (2018).
Culver, J. O., Hull, J. L., Dunne, D. F. & Burke, W. Oncologists’ opinions on genetic testing for breast and ovarian cancer. Genet. Med. 3, 120–125 (2001).
Teng, I. & Spigelman, A. Attitudes and knowledge of medical practitioners to hereditary cancer clinics and cancer genetic testing. Fam. Cancer 13, 311–324 (2014).
Teer, J. K. et al. Evaluating somatic tumor mutation detection without matched normal samples. Hum. Genomics 11, 22 (2017).
Damodaran, S., Berger, M. F. & Roychowdhury, S. Clinical tumor sequencing: opportunities and challenges for precision cancer medicine. Am. Soc. Clin. Oncol. Educ. Book 35, e175–e182 (2015).
Robinson, D. R. et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017).
Van Allen, E. M. et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 20, 682–688 (2014).
Van Allen, E. M. et al. A comparative assessment of clinical whole exome and transcriptome profiling across sequencing centers: implications for precision cancer medicine. Oncotarget 7, 52888–52899 (2016).
Robbe, P. et al. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genet. Med. 20, 1196–1205 (2018).
Staaf, J. et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat. Med. 25, 1526–1533 (2019).
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016).
Zheng, Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 20, 1479–1484 (2014).
Cieslik, M. et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 25, 1372–1381 (2015).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018).
Miller, A. M. et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 565, 654–658 (2019).
Goto, K. et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J. Thorac. Oncol. 7, 115–121 (2012).
Husain, H. et al. Cell-free DNA from ascites and pleural effusions: molecular insights into genomic aberrations and disease biology. Mol. Cancer Ther. 16, 948–955 (2017).
Chang, H. W. et al. Urinary cell-free DNA as a potential tumor marker for bladder cancer. Int. J. Biol. Markers 22, 287–294 (2007).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
Lecomte, T. et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int. J. Cancer 100, 542–548 (2002).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Dawson, S.-J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Garcia-Murillas, I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 7, 302ra133 (2015).
Kinde, I., Wu, J., Papadopoulos, N., Kinzler, K. W. & Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. USA 108, 9530–9535 (2011).
Oxnard, G. R. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 34, 3375–3382 (2016).
Shaw, J. A. et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin. Cancer Res. 23, 88–96 (2017).
Goodall, J. et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 7, 1006–1017 (2017).
Reinert, T. et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131 (2019).
Christensen, E. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
Coombes, R. C. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
Hao, X. et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl Acad. Sci. USA 114, 7414–7419 (2017).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016).
Landau, D. A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Tarabichi, M. et al. A practical guide to cancer subclonal reconstruction from DNA sequencing. Nat. Methods 18, 144–155 (2021).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Voss, M. H. et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin. Cancer Res. 20, 1955–1964 (2014).
Hyman, D. M. et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 554, 189–194 (2018).
Bielski, C. M. et al. Widespread selection for oncogenic mutant allele imbalance in cancer. Cancer Cell 34, 852–862.e4 (2018).
Jones, P. A. & Baylin, S. B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428 (2002).
Vasan, N. et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 366, 714–723 (2019).
Paraiso, K. H. T. et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 71, 2750–2760 (2011).
Xing, F. et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene 31, 446–457 (2012).
Whittaker, S. R. et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 3, 350–362 (2013).
Nissan, M. H. et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 74, 2340–2350 (2014).
Alexandrov, L. B. & Stratton, M. R. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr. Opin. Genet. Dev. 24, 52–60 (2014).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Niu, B. et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30, 1015–1016 (2014).
Escudié, F. et al. MIAmS: microsatellite instability detection on NGS amplicons data. Bioinformatics https://doi.org/10.1093/bioinformatics/btz797 (2019).
Huang, M. N. et al. MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci. Rep. 5, 13321 (2015).
Abida, W. et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 5, 471–478 (2019).
Wu, Y.-M. et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 173, 1770–1782.e14 (2018).
Wang, Y. K. et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat. Genet. 49, 856–865 (2017).
Taylor-Weiner, A. et al. Genomic evolution and chemoresistance in germ-cell tumours. Nature 540, 114–118 (2016).
Angus, L. et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat. Genet. 51, 1450–1458 (2019).
Salgado, R. et al. Addressing the dichotomy between individual and societal approaches to personalised medicine in oncology. Eur. J. Cancer 114, 128–136 (2019).
Salgado, R. et al. How current assay approval policies are leading to unintended imprecision medicine. Lancet Oncol. 21, 1399–1401 (2020).
Rosell, R. et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 13, 239–246 (2012).
Mok, T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361, 947–957 (2009).
Fukuoka, M. et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J. Clin. Oncol. 21, 2237–2246 (2003).
Kris, M. G. et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA 290, 2149–2158 (2003).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004).
Pao, W. et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004).
Hann, C. L. & Brahmer, J. R. ‘Who should receive epidermal growth factor receptor inhibitors for non-small cell lung cancer and when?’. Curr. Treat. Options Oncol. 8, 28–37 (2007).
Soda, M. et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007).
Davies, K. D. et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin. Cancer Res. 18, 4570–4579 (2012).
Takeuchi, K. et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–381 (2012).
Gautschi, O. et al. Targeted therapy for patients with BRAF-mutant lung cancer: results from the European EURAF cohort. J. Thorac. Oncol. 10, 1451–1457 (2015).
Stephens, P. et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 431, 525–526 (2004).
Shimamura, T. et al. Non-small-cell lung cancer and Ba/F3 transformed cells harboring the ERBB2 G776insV_G/C mutation are sensitive to the dual-specific epidermal growth factor receptor and ERBB2 inhibitor HKI-272. Cancer Res. 66, 6487–6491 (2006).
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Paik, P. K. et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 8, 842–849 (2015).
Govindan, R. et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 150, 1121–1134 (2012).
Douillard, J.-Y. et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J. Clin. Oncol. 28, 4697–4705 (2010).
Bokemeyer, C. et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann. Oncol. 22, 1535–1546 (2011).
Joseph, E. W. et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc. Natl Acad. Sci. USA 107, 14903–14908 (2010).
Su, F. et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 366, 207–215 (2012).
Callahan, M. K. et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N. Engl. J. Med. 367, 2316–2321 (2012).
Sanchez-Laorden, B. et al. BRAF inhibitors induce metastasis in RAS mutant or inhibitor-resistant melanoma cells by reactivating MEK and ERK signaling. Sci. Signal. 7, ra30 (2014).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 (2019).
Marabelle, A. et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 21, 1353–1365 (2020).
Hellmann, M. D. et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 33, 853–861.e4 (2018).
Marabelle, A. et al. 1192O - Association of tumour mutational burden with outcomes in patients with select advanced solid tumours treated with pembrolizumab in KEYNOTE-158. Ann. Oncol. 30, v477–v478 (2019).
Graff, J. N. et al. A phase II single-arm study of pembrolizumab with enzalutamide in men with metastatic castration-resistant prostate cancer progressing on enzalutamide alone. J Immunother. Cancer 8, e000642 (2020).
O’Reilly, E. M. et al. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol. 5, 1431–1438 (2019).
Spranger, S., Bao, R. & Gajewski, T. F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015).
Xiao, Q. et al. DKK2 imparts tumor immunity evasion through β-catenin-independent suppression of cytotoxic immune-cell activation. Nat. Med. 24, 262–270 (2018).
Harding, J. J. et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin. Cancer Res. 25, 2116–2126 (2019).
Peters, S. et al. Phase II trial of atezolizumab as first-line or subsequent therapy for patients with programmed death-ligand 1-selected advanced non–small-cell lung cancer (BIRCH). J. Clin. Oncol. 35, 2781–2789 (2017).
Haratani, K. et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann. Oncol. 28, 1532–1539 (2017).
Chowell, D. et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
D’Angelo, S. P. et al. Avelumab in patients with previously treated metastatic Merkel cell carcinoma: long-term data and biomarker analyses from the single-arm phase 2 JAVELIN Merkel 200 trial. J. Immunother. Cancer 8, e000674 (2020).
Janjigian, Y. Y. et al. Genetic predictors of response to systemic therapy in esophagogastric cancer. Cancer Discov. 8, 49–58 (2018).
Kim, S. T. et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 24, 1449–1458 (2018).
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019).
Wang, R. F. & Rosenberg, S. A. Human tumor antigens for cancer vaccine development. Immunol. Rev. 170, 85–100 (1999).
Acknowledgements
The authors thank M. F. Berger, H. Al-Ahmadie, C. Ho, S. Sethi, A. Zehir and S. Nandakumar for their invaluable contributions to the figures. They are also grateful to the Molecular Diagnostics Service and the OncoKB team, particularly M. Ladanyi, H. Zhang and R. Kundra, for their assistance and insightful suggestions.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
D.B.S. serves on the Scientific Advisory Board for Loxo Oncology at Eli Lilly, Pfizer, Scorpion Therapeutics, BridgeBio and Vividion Therapeutics, owned stock at Loxo Oncology at Eli Lilly and Scorpion Therapeutics, and received honoraria from Illumina and Eli Lilly. D.C. declares no competing interests.
Additional information
Peer review information
Nature Reviews Genetics thanks J. Martens and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Precision oncology
-
The process of using molecular data from the analysis of a patient’s tumour or healthy cells to inform treatment selection.
- Companion diagnostics
-
Within the context of precision oncology, companion diagnostics are medical devices designed to identify the subset of patients most likely to respond to and benefit from a targeted or other systemic or local therapy.
- Next-generation sequencing
-
(NGS). Massively parallel high-throughput sequencing methods designed to analyse DNA or RNA more rapidly and at higher resolution than older methods such as Sanger sequencing.
- Cell-free DNA
-
(cfDNA). In the context of this article, circulating tumour DNA, that is, DNA fragments shed by the tumour into the blood.
- Basket trials
-
A clinical trial design that prospectively accrues patients with a specific molecular alteration irrespective of tumour type.
- Mutational signatures
-
Patterns of base pair substitutions or structural abnormalities that are often characteristic of exogenous or endogenous mutational processes (such as tobacco smoking or DNA repair pathway mutations).
- Somatic mutations
-
Non-heritable mutations that may arise in any cell except germ cells (sperm or ova). Although somatic mutations have classically been defined as those found specifically in tumour but not in healthy cells, accumulation of somatic mutations is common in non-transformed, histologically normal-appearing cells as patients age.
- Drivers
-
Mutations that enhance tumour cell fitness by providing a growth or survival advantage.
- Passengers
-
Biologically inert mutations with no impact on tumour cell fitness.
- Clinically actionable mutations
-
A subset of driver mutations that are predictive biomarkers of drug response or resistance.
- Synthetic lethal mechanism
-
An interaction between two genes whereby loss of function of both (due to mutation, epigenetic silencing or drug inhibition) results in tumour cell death, whereas loss of function or inhibition of either individual gene does not.
- Mutational hotspots
-
Mutations identified in a population of patients with cancer more frequently than expected by chance.
- Germline variants
-
Heritable mutations that were present in germ cells and consequently found in all cells of the descendants.
- Resistance mutations
-
Mutations that increase tumour cell fitness under the selective pressure of a systemic therapy.
- Penetrance
-
A measure of the proportion of individuals with a mutant allele in a defined population who manifest the associated phenotype. For germline variants associated with increased cancer predisposition, the penetrance is the proportion of patients who develop the associated cancer type.
- Whole-exome sequencing
-
(WES). Sequencing of all protein-coding regions (or exons) in the genome.
- Whole-genome sequencing
-
(WGS). Sequencing of the entire genome including non-coding sequences.
- Clonal haematopoiesis
-
The acquisition of somatic genomic alterations in haematopoietic stem and/or progenitor cells, resulting in clonal expansion.
- Capture-based DNA sequencing
-
A method for selectively sequencing targeted regions of the genome using baits that hybridize with specific regions of DNA.
- Clonal mutations
-
Mutations present in all of a patient’s cancer cells.
- Subclonal mutations
-
Mutations present in only a fraction of a patient’s cancer cells.
- Cancer cell fraction
-
The estimated fraction of cancer cells that harbour a specific mutation.
- Variant allele frequency
-
The fraction of mutant versus total sequencing reads at the mutation locus.
- Allelic configuration
-
The number of mutant and wild-type alleles, which because of copy number gain or loss can be less than or greater than two.
- Knudsen’s two-hit hypothesis
-
The hypothesis, proposed by Alfred Knudsen in 1971, that for tumour suppressor genes that are recessive in nature, in order for a phenotype to manifest, both alleles must be inactivated (biallelic inactivation). This may be achieved through multiple mechanisms including deletions (either chromosomal or subchromosomal), epigenetic silencing or mutation.
- Loss of heterozygosity
-
A common form of allelic imbalance in which a heterozygous somatic alteration becomes homozygous following loss of the wild-type allele.
- Clonal fitness
-
The relative growth, survival or metastatic potential advantage of a cancer cell clone over other cancer cells within the tumour or non-cancer cells. The term fitness here derives its origins from the concept of natural selection in evolutionary biology.
- Integer copy number
-
The copy number of a gene or localized DNA segment represented as an integer value. For missense mutations, the number of mutated and wild-type alleles in the cell.
Rights and permissions
About this article
Cite this article
Chakravarty, D., Solit, D.B. Clinical cancer genomic profiling. Nat Rev Genet 22, 483–501 (2021). https://doi.org/10.1038/s41576-021-00338-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-021-00338-8
This article is cited by
-
m6A modification patterns are associated with copy number burden and tumor immune landscape in thyroid cancer
BMC Endocrine Disorders (2023)
-
A novel necroptosis signature for predicting survival in lung adenocarcinoma
BMC Medical Genomics (2023)
-
SNPeffect 5.0: large-scale structural phenotyping of protein coding variants extracted from next-generation sequencing data using AlphaFold models
BMC Bioinformatics (2023)
-
Comprehensive analysis of PILRΑ’s association with the prognosis, tumor immune infiltration, and immunotherapy in pan-cancer
Scientific Reports (2023)
-
TEQUILA-seq: a versatile and low-cost method for targeted long-read RNA sequencing
Nature Communications (2023)