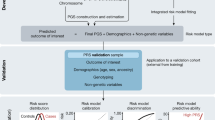
Abstract
Accurate prediction of disease risk based on the genetic make-up of an individual is essential for effective prevention and personalized treatment. Nevertheless, to date, individual genetic variants from genome-wide association studies have achieved only moderate prediction of disease risk. The aggregation of genetic variants under a polygenic model shows promising improvements in prediction accuracies. Increasingly, electronic health records (EHRs) are being linked to patient genetic data in biobanks, which provides new opportunities for developing and applying polygenic risk scores in the clinic, to systematically examine and evaluate patient susceptibilities to disease. However, the heterogeneous nature of EHR data brings forth many practical challenges along every step of designing and implementing risk prediction strategies. In this Review, we present the unique considerations for using genotype and phenotype data from biobank-linked EHRs for polygenic risk prediction.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Preiss, D. & Kristensen, S. L. The new pooled cohort equations risk calculator. Can. J. Cardiol. 31, 613–619 (2015).
Antoniou, A. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 72, 1117–1130 (2003).
O’Sullivan, B. P. & Freedman, S. D. Cystic fibrosis. Lancet 373, 1891–1904 (2009).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Lo, A., Chernoff, H., Zheng, T. & Lo, S.-H. Why significant variables aren’t automatically good predictors. Proc. Natl Acad. Sci. USA 112, 13892–13897 (2015).
Visscher, P. M. et al. 10 Years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Bogdan, R., Baranger, D. A. A. & Agrawal, A. Polygenic risk scores in clinical psychology: bridging genomic risk to individual differences. Annu. Rev. Clin. Psychol. 14, 119–157 (2018).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Zhang, J.-P. et al. Schizophrenia polygenic risk score as a predictor of antipsychotic efficacy in first-episode psychosis. Am. J. Psychiatry 176, 21–28 (2019).
Jones, H. J. et al. Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiatry 73, 221 (2016).
Kohane, I. S. Using electronic health records to drive discovery in disease genomics. Nat. Rev. Genet.12, 417–428 (2011).
Fritsche, L. G. et al. Association of polygenic risk scores for multiple cancers in a phenome-wide study: results from the michigan genomics initiative. Am. J. Hum. Genet. 102, 1048–1061 (2018). This analysis uses biobank-linked EHR data to study PRS associations with cancers.
Banda, Y. et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics 200, 1285–1295 (2015).
Kvale, M. N. et al. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics 200, 1051–1060 (2015).
Li, R. et al. A regression framework to uncover pleiotropy in large-scale electronic health record data. J. Am. Med. Informatics Assoc 26, 1083–1090 (2019).
McCarty, C. A., Wilke, R. A., Giampietro, P. F., Wesbrook, S. D. & Caldwell, M. D. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Per. Med. 2, 49–79 (2005).
Nagai, A. et al. Overview of the Biobank Japan project: study design and profile. J. Epidemiol. 27, S2–S8 (2017).
Cho, S. Y. et al. Opening of the National Biobank of Korea as the infrastructure of future biomedical science in Korea. Osong Public. Heal. Res. Perspect. 3, 177–184 (2012).
Chen, Z. et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int. J. Epidemiol. 40, 1652–1666 (2011).
Locke, A. E. et al. Exome sequencing of Finnish isolates enhances rare-variant association power. Nature 572, 323–328 (2019).
Sankar, P. L. & Parker, L. S. The Precision Medicine Initiative’s all of us research program: an agenda for research on its ethical, legal, and social issues. Genet. Med. 19, 743–750 (2017).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). This paper presents one of the largest genetic-linked patient clinical data sets that is publicly available to researchers.
Casey, J. A., Schwartz, B. S., Stewart, W. F. & Adler, N. E. Using electronic health records for population health research: a review of methods and applications. Annu. Rev. Public Health 37, 61–81 (2016).
Chatterjee, N., Shi, J. & García-Closas, M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 17, 392–406 (2016). This review article provides an overview of risk prediction methods and approaches to incorporate a PRS into risk models.
Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018). This paper presents a review of the background of the PRS and how it can be utilized for risk predictions.
Li, R., Chen, Y. & Moore, J. H. Integration of genetic and clinical information to improve imputation of data missing from electronic health records. J. Am. Med. Informatics Assoc. 26, 1056–1063 (2019).
Shi, J. et al. Winner’s curse correction and variable thresholding improve performance of polygenic risk modeling based on genome-wide association study summary-level data. PLoS Genet.12, e1006493 (2016).
Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015). This study shows that the accuracy of a PRS is affected by the modelling of linkage disequilibrium between SNPs.
Mak, T. S. H., Porsch, R. M., Choi, S. W., Zhou, X. & Sham, P. C. Polygenic scores via penalized regression on summary statistics. Genet. Epidemiol. 41, 469–480 (2017).
Choi, S. W., Mak, T. S. H. & O’Reilly, P. F. A guide to performing polygenic risk score analyses. Preprint at bioRxiv https://doi.org/10.1101/416545 (2018).
Fritsche, L. G. et al. Exploring various polygenic risk scores for skin cancer in the phenomes of the Michigan Genomics Initiative and the UK Biobank with a visual catalog: PRSWeb. PLoS Genet. 15, e1008202 (2019).
Reus, L. M. et al. Association of polygenic risk for major psychiatric illness with subcortical volumes and white matter integrity in UK Biobank. Sci. Rep. 7, 42140 (2017).
Mavaddat, N. et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 104, 21–34 (2019).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018). This study demonstrates that a PRS can identify individuals who have a clinically significantly increased risk of coronary artery disease, atrial fibrillation, T2DM, inflammatory bowel disease and breast cancer.
Khera, A. V. et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell 177, 587–596.e9 (2019).
Polubriaginof, F. C. G. et al. Disease heritability inferred from familial relationships reported in medical records. Cell 173, 1692–1704.e11 (2018).
DeBoever, C. et al. Assessing digital phenotyping to enhance genetic studies of human diseases. Preprint at bioRxiv https://doi.org/10.1101/738856 (2019).
Robinson, J. R., Wei, W.-Q., Roden, D. M. & Denny, J. C. Defining phenotypes from clinical data to drive genomic research. Annu. Rev. Biomed. Data Sci. 1, 69–92 (2018).
Wei, W.-Q. & Denny, J. C. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 7, 41 (2015).
Denny, J. C. et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics 26, 1205–1210 (2010).
Chiu, P.-H. & Hripcsak, G. EHR-based phenotyping: bulk learning and evaluation. J. Biomed. Inform. 70, 35–51 (2017).
Banda, J. M., Seneviratne, M., Hernandez-Boussard, T. & Shah, N. H. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu. Rev. Biomed. Data Sci. 1, 53–68 (2018).
Ritchie, M. D. Large-scale analysis of genetic and clinical patient data. Annu. Rev. Biomed. Data Sci. 1, 263–274 (2018).
Hripcsak, G. & Albers, D. J. Next-generation phenotyping of electronic health records. J. Am. Med. Inform. Assoc. 20, 117–121 (2013).
Kirby, J. C. et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J. Am. Med. Inform. Assoc. 23, 1046–1052 (2016). This paper discusses PheKB, which contains a wide range of phenotyping algorithms that can automatically extract phenotypes from EHR data.
Liao, K. P. et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 350, h1885 (2015).
Yu, S. et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J. Am. Med. Inform. Assoc. 24, e143–e149 (2017).
Beaulieu-Jones, B. K. et al. Characterizing and managing missing structured data in electronic health records: data analysis. JMIR Med. Inform. 6, e11 (2018).
Kleinsinger, F. The unmet challenge of medication nonadherence. Perm. J. 22, 18-033 (2018).
Kho, A. N. et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J. Am. Med. Inform. Assoc. 19, 212–218 (2012).
Peissig, P. L. et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J. Am. Med. Inform. Assoc. 19, 225–234 (2012).
Halpern, Y., Horng, S., Choi, Y. & Sontag, D. Electronic medical record phenotyping using the anchor and learn framework. J. Am. Med. Inform. Assoc. 23, 731–740 (2016).
Dumitrescu, L. et al. Genome-wide study of resistant hypertension identified from electronic health records. PLoS One 12, e0171745 (2017).
Crosslin, D. R. et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum. Genet. 131, 639–652 (2012).
Choquet, H. et al. A large multi-ethnic genome-wide association study identifies novel genetic loci for intraocular pressure. Nat. Commun. 8, 2108 (2017).
Gudbjartsson, D. F. et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 47, 435–444 (2015).
Robinson, P. N. et al. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83, 610–615 (2008).
Randorff Højen, A. & Rosenbeck Gøeg, K. SNOMED CT implementation. Methods Inf. Med. 51, 529–538 (2012).
Vreeman, D. J., McDonald, C. J. & Huff, S. M. LOINC®: a universal catalogue of individual clinical observations and uniform representation of enumerated collections. Int. J. Funct. Inform. Personal. Med. 3, 273 (2010).
Schulam, P., Wigley, F. & Saria, S. Clustering longitudinal clinical marker trajectories from electronic health data: applications to phenotyping and endotype discovery. Proc. Natl Conf. Artif. Intell. 4, 2956–2964 (2015).
Duan, R. et al. An empirical study for impacts of measurement errors on EHR based association studies. AMIA Annu. Symp. Proc. 2016, 1764–1773 (2017).
Chen, Y., Wang, J., Chubak, J. & Hubbard, R. A. Inflation of type I error rates due to differential misclassification in EHR-derived outcomes: empirical illustration using breast cancer recurrence. Pharmacoepidemiol. Drug Saf. 28, 264–268 (2019).
Li, R., Tong, J., Duan, R., Chen, Y. & Moore, J. H. Evaluation of phenotyping errors on polygenic risk score predictions. Proc. Int. Joint Conf. Biomed. Eng. Syst. Technol. https://doi.org/10.5220/0008935301230130 (2020).
Wells, B. J., Chagin, K. M., Nowacki, A. S. & Kattan, M. W. Strategies for handling missing data in electronic health record derived data. EGEMS 1, 1035 (2013).
Zheng, T. et al. A machine learning-based framework to identify type 2 diabetes through electronic health records. Int. J. Med. Inform. 97, 120–127 (2017).
Gustafson, E., Pacheco, J., Wehbe, F., Silverberg, J. & Thompson, W. A machine learning algorithm for identifying atopic dermatitis in adults from electronic health records. IEEE Int. Conf. Healthc. Inform. 2017, 83–90 (2017).
Zhou, S.-M. et al. Defining disease phenotypes in primary care electronic health records by a machine learning approach: a case study in identifying rheumatoid arthritis. PLoS One 11, e0154515 (2016).
Carroll, R. J., Eyler, A. E. & Denny, J. C. Naïve electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu. Symp. Proc. 2011, 189–196 (2011).
Cimino, J. J., Lancaster, W. J. & Wyatt, M. C. Classification of clinical research study eligibility criteria to support multi-stage cohort identification using clinical data repositories. Stud. Health Technol. Inform. 245, 341–345 (2017).
Gottesman, O. et al. The electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 15, 761–771 (2013).
Zhao, J. et al. Learning from longitudinal data in electronic health record and genetic data to improve cardiovascular event prediction. Sci. Rep. 9, 717 (2019).
Martin, A. R. et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 100, 635–649 (2017).
Sohail, M. et al. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife 8, 39702 (2019).
Zeng, Z., Deng, Y., Li, X., Naumann, T. & Luo, Y. Natural language processing for EHR-based computational phenotyping. IEEE/ACM Trans. Comput. Biol. Bioinform. 16, 139–153 (2019).
Denaxas, S. et al. Methods for enhancing the reproducibility of biomedical research findings using electronic health records. BioData Min. 10, 31 (2017).
Berg, J. J. et al. Reduced signal for polygenic adaptation of height in UK Biobank. eLife 8, e39725 (2019).
Gao, X. R., Huang, H. & Kim, H. Polygenic risk score is associated with intraocular pressure and improves glaucoma prediction in the UK Biobank cohort. Transl. Vis. Sci. Technol. 8, 10 (2019).
Stang, P. E. et al. Advancing the science for active surveillance: rationale and design for the observational medical outcomes partnership. Ann. Intern. Med. 153, 600 (2010).
Hripcsak, G. et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud. Health Technol. Inform. 216, 574–578 (2015).
Duan, R., Boland, M. R., Moore, J. H. & Chen, Y. ODAL: a one-shot distributed algorithm to perform logistic regressions on electronic health records data from multiple clinical sites. Pac. Symp. Biocomput. 24, 30–41 (2019).
Duan, R. et al. Learning from electronic health records across multiple sites: a communication-efficient and privacy-preserving distributed algorithm. J. Am. Med. Informatics Assoc. 27, 376–385 (2019).
Ohno-Machado, L., Kim, J., Gabriel, R. A., Kuo, G. M. & Hogarth, M. A. Genomics and electronic health record systems. Hum. Mol. Genet. 27, R48–R55 (2018).
Farmer, R. et al. Promises and pitfalls of electronic health record analysis. Diabetologia 61, 1241–1248 (2018).
Denny, J. C. et al. The “All of Us” research program. N. Engl. J. Med. 381, 668–676 (2019).
Coloma, P. M. et al. Combining electronic healthcare databases in Europe to allow for large-scale drug safety monitoring: the EU-ADR project. Pharmacoepidemiol. Drug Saf. 20, 1–11 (2011).
Trifiro, G. et al. The EU-ADR project: preliminary results and perspective. Stud. Health Technol. Inform. 148, 43–49 (2009).
Lai, E. C.-C. et al. Applying a common data model to Asian databases for multinational pharmacoepidemiologic studies: opportunities and challenges. Clin. Epidemiol. 10, 875–885 (2018).
Platt, R. W. et al. How pharmacoepidemiology networks can manage distributed analyses to improve replicability and transparency and minimize bias. Pharmacoepidemiol. Drug Saf. 29, 3–7 (2019).
Greco, T., Zangrillo, A., Biondi-Zoccai, G. & Landoni, G. Meta-analysis: pitfalls and hints. Heart Lung Vessel. 5, 219–225 (2013).
Lu, C.-L. et al. WebDISCO: a web service for distributed Cox model learning without patient-level data sharing. J. Am. Med. Inform. Assoc. 22, ocv083 (2015).
Wu, Y., Jiang, X., Kim, J. & Ohno-Machado, L. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. J. Am. Med. Inform. Assoc. 19, 758–764 (2012).
Yixin Chen et al. Regression cubes with lossless compression and aggregation. IEEE Trans. Knowl. Data Eng. 18, 1585–1599 (2006).
Wang, J., Kolar, M., Srebro, N. & Zhang, T. Efficient distributed learning with sparsity. Preprint at arXiv https://arxiv.org/abs/1605.07991 (2016).
Wray, N. R., Kemper, K. E., Hayes, B. J., Goddard, M. E. & Visscher, P. M. Complex trait prediction from genome data: contrasting EBV in livestock to PRS in humans. Genetics 211, 1131–1141 (2019).
Powers, D. M. W. Evaluation: from precision, recall and F-factor to ROC, informedness, markedness & correlation. J. Mach. Learn. Technol. 2, 37–63 (2011).
Choudhury, P. P. et al. iCARE: an R package to build, validate and apply absolute risk models. PLoS One 15, e0228198 (2020).
Choudhury, P. P. et al. Comparative validation of breast cancer risk prediction models and projections for future risk stratification. J. Natl. Cancer Inst. 112, 278–285 (2019).
Violán, C. et al. Comparison of the information provided by electronic health records data and a population health survey to estimate prevalence of selected health conditions and multimorbidity. BMC Public Health 13, 251 (2013).
Price, W. N. & Cohen, I. G. Privacy in the age of medical big data. Nat. Med. 25, 37–43 (2019).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Rammos, A., Gonzalez, L. A. N., Weinberger, D. R., Mitchell, K. J. & Nicodemus, K. K. The role of polygenic risk score gene-set analysis in the context of the omnigenic model of schizophrenia. Neuropsychopharmacology 44, 1562–1569 (2019).
Meisner, A., Kundu, P. & Chatterjee, N. Case-only analysis of gene–environment interactions using polygenic risk scores. Am. J. Epidemiol. 188, 2013–2020 (2019).
Manolio, T. A. Using the data we have: improving diversity in genomic research. Am. J. Hum. Genet. 105, 233–236 (2019).
Khoury, M. J. & Mensah, G. A. Is it time to integrate polygenic risk scores into clinical practice? Let’s do the science first and follow the evidence wherever it takes us! CDC https://blogs.cdc.gov/genomics/2019/06/03/is-it-time/ (2019)
Gibson, G. On the utilization of polygenic risk scores for therapeutic targeting. PLoS Genet. 15, e1008060 (2019).
Lee, A. et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 21, 1708–1718 (2019). This study integrates known cancer genes, a PRS, lifestyle risk factors and mammographic density to better estimate breast cancer risk in women.
Pashayan, N. et al. Reducing overdiagnosis by polygenic risk-stratified screening: findings from the Finnish section of the ERSPC. Br. J. Cancer 113, 1086–1093 (2015).
Lambert, S. A., Abraham, G. & Inouye, M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 28, R133–R142 (2019).
Arnett, D. K. et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 74, e177–e232 (2019).
Bielinski, S. J. & Pathak, J. Heart failure with differentiation between reduced and preserved ejection fraction — Phenotype algorithm pseudo code (Mayo Clinic). NCBI https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/GetPdf.cgi?id=phd004988.1 (2014)
National Center for Health Statistics & Centers for Disease Control and Prevention. International classification of diseases, ninth revision (ICD-9) (CDC, 1998).
Côté, R. A. & Robboy, S. Progress in medical information management. JAMA 243, 756 (1980).
McDonald, C. J. et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin. Chem. 49, 624–633 (2003).
Fung, K. W., McDonald, C. & Bray, B. E. RxTerms — a drug interface terminology derived from RxNorm. AMIA Annu. Symp. Proc. 2008, 227–231 (2008).
ICD.Codes. The switch from ICD-9 to ICD-10: when and why. ICD.Codes https://icd.codes/articles/icd9-to-icd10-explained (2015)
Topaz, M., Shafran-Topaz, L. & Bowles, K. H. ICD-9 to ICD-10: evolution, revolution, and current debates in the United States. Perspect. Heal. Inf. Manag. 10, 1d (2013).
American Medical Association. Preparing for the ICD-10 code set: the differences between ICD-9 and ICD-10 (AMA, 2014)
Hong, E. P. & Park, J. W. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 10, 117–122 (2012).
Mitt, M. et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet. 25, 869–876 (2017).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019). This study demonstrates the lack of transferability of a PRS across different populations.
Márquez-Luna, C., Loh, P.-R., South Asian Type 2 Diabetes (SAT2D) Consortium, SIGMA Type 2 Diabetes Consortium & Price, A. L. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol. 41, 811–823 (2017). This study uses data from multiple populations to derive a more generalizable PRS for T2DM.
Acknowledgements
This work was supported by National Institutes of Health grants LM010098 and AI116794.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Michigan Genomics Initiative: https://precisionhealth.umich.edu/michigangenomics/
Office of the National Coordinator for Health Information Technology: https://dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php
Phenotype KnowledgeBase (PheKB): https://www.phekb.org/
Precision Medicine Initiative: https://ghr.nlm.nih.gov/primer/precisionmedicine/initiative
Glossary
- Genome-wide association studies
-
(GWAS). Studies in which associations between genetic variation and a phenotype or trait of interest are identified by genotyping cases (for example, diseased individuals) and controls (for example, healthy individuals) for a set of genetic variants that capture variation across the entire genome.
- Polygenic risk score
-
(PRS). A weighted score calculated from numerous genetic variants for predicting disease risk. A PRS is calculated as the sum of risk alleles multiplied by their association coefficients.
- Electronic health records
-
(EHRs). Also known as electronic medical records. Digitally stored patients’ medical history.
- Biobanks
-
Repositories that store biological samples, including blood or tissue samples, for research use. Increasingly, the term biobank is used to denote a population cohort study with stored biological samples.
- Endophenotypes
-
The physiological traits that are related to a disease trait; for example, for hypertension this could include blood pressure, angiotensin levels or salt sensitivity.
- Pleiotropic
-
Pertaining to a gene that affects multiple phenotypes or traits.
- Positive predictive value
-
The proportion of true positives among positive results.
- k-Nearest neighbour
-
A machine learning method that is based on similarities between samples.
- Decision tree
-
A machine learning method that learns decision rules from the data and represents them in a tree-like structure. The tree is used to perform classification or regression.
- Random forest
-
An ensemble approach that learns from multiple decision trees.
- Support vector machine
-
A supervised machine learning method that uses a hyperplane to perform classification.
- Naive Bayes
-
A simple classification method that is based on the Bayes’ theorem.
- Lasso logistic regression
-
A penalized version of regular logistic regression. The additional penalty term forces some features to have zero coefficients.
- Population stratification
-
The presence of allele frequency differences between subpopulations within a larger population.
- Relative risk
-
The ratio of the probability of an event (such as disease) occurring in an at-risk group to the probability of it occurring in a population that is not considered at risk.
- Absolute risk
-
The actual probability of disease occurrence.
- Omnigenic model
-
A model that proposes that the genetic architecture of a trait or disease is affected by many genes.
Rights and permissions
About this article
Cite this article
Li, R., Chen, Y., Ritchie, M.D. et al. Electronic health records and polygenic risk scores for predicting disease risk. Nat Rev Genet 21, 493–502 (2020). https://doi.org/10.1038/s41576-020-0224-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-020-0224-1
This article is cited by
-
An ensemble penalized regression method for multi-ancestry polygenic risk prediction
Nature Communications (2024)
-
Machine learning for the prediction of sepsis-related death: a systematic review and meta-analysis
BMC Medical Informatics and Decision Making (2023)
-
A machine learning model identifies patients in need of autoimmune disease testing using electronic health records
Nature Communications (2023)
-
Polygenic scoring accuracy varies across the genetic ancestry continuum
Nature (2023)
-
Identifying Safety Subgroups at Risk: Assessing the Agreement Between Statistical Alerting and Patient Subgroup Risk
Drug Safety (2023)