Abstract
Cancers and developmental disorders are associated with alterations in the 3D genome architecture in space and time (the fourth dimension). Mammalian 3D genome organization is complex and dynamic and plays an essential role in regulating gene expression and cellular function. To study the causal relationship between genome function and its spatio-temporal organization in the nucleus, new technologies for engineering and manipulating the 3D organization of the genome have been developed. In particular, CRISPR–Cas technologies allow programmable manipulation at specific genomic loci, enabling unparalleled opportunities in this emerging field of 3D genome engineering. We review advances in mammalian 3D genome engineering with a focus on recent manipulative technologies using CRISPR–Cas and related technologies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bickmore, W. A. The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14, 67–84 (2013).
Clowney, E. J. et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151, 724–737 (2012).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Yu, M. & Ren, B. The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 33, 265–289 (2017).
Zheng, H. & Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 20, 535–550 (2019).
Schoenfelder, S. & Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Marchal, C., Sima, J. & Gilbert, D. M. Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-019-0162-y (2019).
Cremer, M. & Cremer, T. Nuclear compartmentalization, dynamics, and function of regulatory DNA sequences. Genes Chromosomes Cancer 58, 427–436 (2019).
Kumaran, R. I., Thakar, R. & Spector, D. L. Chromatin dynamics and gene positioning. Cell 132, 929–934 (2008).
Vermunt, M. W., Zhang, D. & Blobel, G. A. The interdependence of gene-regulatory elements and the 3D genome. J. Cell Biol. 218, 12–26 (2019).
Pombo, A. & Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015).
Anania, C. & Lupianez, D. G. Order and disorder: abnormal 3D chromatin organization in human disease. Brief. Funct. Genomics 19, 128–138 (2020).
Chakraborty, A. & Ay, F. The role of 3D genome organization in disease: from compartments to single nucleotides. Semin. Cell Dev. Biol. 90, 104–113 (2019).
Kim, Y., Zheng, X. & Zheng, Y. Role of lamins in 3D genome organization and global gene expression. Nucleus 10, 33–41 (2019).
Worman, H. J., Fong, L. G., Muchir, A. & Young, S. G. Laminopathies and the long strange trip from basic cell biology to therapy. J. Clin. Invest. 119, 1825–1836 (2009).
Lupianez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). This work shows that TAD disruptions are associated with human limb malformations, and it builds mouse models of these diseases by CRISPR editing of TAD boundary elements.
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Guo, Y. et al. CRISPR-mediated deletion of prostate cancer risk-associated CTCF loop anchors identifies repressive chromatin loops. Genome Biol. 19, 160 (2018).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Valton, A. L. & Dekker, J. TAD disruption as oncogenic driver. Curr. Opin. Genet. Dev. 36, 34–40 (2016).
Stadhouders, R., Filion, G. J. & Graf, T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569, 345–354 (2019).
Peric-Hupkes, D. et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 (2010).
Williams, R. R. et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119, 132–140 (2006).
Hiratani, I. et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6, e245 (2008).
van Steensel, B. & Belmont, A. S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017).
Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 (2002).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384 (2016).
Beagan, J. A. et al. Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression. Nat. Neurosci. 23, 707–717 (2020).
Kempfer, R. & Pombo, A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 21, 207–226 (2020).
Schmitt, A. D., Hu, M. & Ren, B. Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell Biol. 17, 743–755 (2016).
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Zheng, M. et al. Multiplex chromatin interactions with single-molecule precision. Nature 566, 558–562 (2019).
Fullwood, M. J. et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 (2009).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Langer-Safer, P. R., Levine, M. & Ward, D. C. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA 79, 4381–4385 (1982).
Denker, A. & de Laat, W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 30, 1357–1382 (2016).
Rowley, M. J. & Corces, V. G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800 (2018).
van Steensel, B. & Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18, 424–428 (2000).
Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757 (2018).
Engreitz, J. M., Ollikainen, N. & Guttman, M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 (2016).
Boettiger, A. N. et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016).
Nir, G. et al. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 14, e1007872 (2018).
Tan, L., Xing, D., Chang, C. H., Li, H. & Xie, X. S. Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018).
Nguyen, H. Q. et al. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat. Methods 17, 822–832 (2020).
Su, J. H., Zheng, P., Kinrot, S. S., Bintu, B. & Zhuang, X. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182, 1641–1659 (2020).
Mateo, L. J. et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376 (2018).
Ma, H. et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 34, 528–530 (2016).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Dominguez, A. A., Lim, W. A. & Qi, L. S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 17, 5–15 (2016).
Wang, W., Zhang, L., Wang, X. & Zeng, Y. The advances in CRISPR technology and 3D genome. Semin. Cell Dev. Biol. 90, 54–61 (2019).
Wang, H., La Russa, M. & Qi, L. S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264 (2016).
Morgan, S. L. et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 8, 15993 (2017). This study develops a programmable, chemical-inducible CLOuD9 technique to induce loop formation between two genomic loci targeted by orthogonal dCas9 proteins, which can induce loop formation to regulate gene expression.
Wang, H. et al. CRISPR-mediated programmable 3D genome positioning and nuclear organization. Cell 175, 1405–1417 (2018). This article describes a CRISPR-GO technique to programmably target dCas9-bound genomic loci to the nuclear periphery and to mediate interactions between target genomic loci and different nuclear bodies.
Liu, X. S. et al. Editing DNA methylation in the mammalian genome. Cell 167, 233–247 (2016).
Knott, G. J. & Doudna, J. A. CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018).
Robinett, C. C. et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135, 1685–1700 (1996).
Kim, Y. G., Cha, J. & Chandrasegaran, S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA 93, 1156–1160 (1996).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Tan, L., Xing, D., Daley, N. & Xie, X. S. Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nat. Struct. Mol. Biol. 26, 297–307 (2019).
Cremer, T. & Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010).
Mao, Y. S., Zhang, B. & Spector, D. L. Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 (2011).
Sawyer, I. A., Sturgill, D. & Dundr, M. Membraneless nuclear organelles and the search for phases within phases. Wiley Interdiscip. Rev. RNA 10, e1514 (2019).
Guelen, L. et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008).
Lochs, S. J. A., Kefalopoulou, S. & Kind, J. Lamina associated domains and gene regulation in development and cancer. Cells https://doi.org/10.3390/cells8030271 (2019).
van de Corput, M. P. et al. Super-resolution imaging reveals three-dimensional folding dynamics of the beta-globin locus upon gene activation. J. Cell Sci. 125, 4630–4639 (2012).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Hansen, A. S. CTCF as a boundary factor for cohesin-mediated loop extrusion: evidence for a multi-step mechanism. Nucleus 11, 132–148 (2020).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, E6456–E6465 (2015).
Ganji, M. et al. Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 (2018).
Alipour, E. & Marko, J. F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 40, 11202–11212 (2012).
Nasmyth, K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673–745 (2001).
Ong, C. T. & Corces, V. G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014).
Pugacheva, E. M. et al. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl Acad. Sci. USA 117, 2020–2031 (2020).
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S. & Gregory, P. D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Christian, M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010).
Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Ishihara, K., Nakamoto, M. & Nakao, M. DNA methylation-independent removable insulator controls chromatin remodeling at the HOXA locus via retinoic acid signaling. Hum. Mol. Genet. 25, 5383–5394 (2016).
Darrow, E. M. et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl Acad. Sci. USA 113, E4504–E4512 (2016).
Guo, Y. et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015).
Giorgetti, L. et al. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579 (2016).
Narendra, V. et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015).
Bonora, G. et al. Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome. Nat. Commun. 9, 1445 (2018).
de Wit, E. et al. CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015).
Barutcu, A. R., Maass, P. G., Lewandowski, J. P., Weiner, C. L. & Rinn, J. L. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun. 9, 1444 (2018).
Williamson, I. et al. Developmentally regulated Shh expression is robust to TAD perturbations. Development https://doi.org/10.1242/dev.179523 (2019).
Li, J. et al. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J. Mol. Cell Biol. 7, 284–298 (2015).
Zhang, D. et al. Alteration of genome folding via contact domain boundary insertion. Nat. Genet. 52, 1076–1087 (2020).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588 (2017).
Sima, J. et al. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell 176, 816–830 (2019).
Gassler, J. et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36, 3600–3618 (2017).
Busslinger, G. A. et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017).
Haarhuis, J. H. I. et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707 (2017).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320 (2017).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944 (2017).
Hyle, J. et al. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer-promoter looping. Nucleic Acids Res. 47, 6699–6713 (2019).
Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017).
Thiecke, M. J. et al. Cohesin-dependent and -independent mechanisms mediate chromosomal contacts between promoters and enhancers. Cell Rep. 32, 107929 (2020).
Natsume, T., Kiyomitsu, T., Saga, Y. & Kanemaki, M. T. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Zuin, J. et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl Acad. Sci. USA 111, 996–1001 (2014).
Kadauke, S. & Blobel, G. A. Chromatin loops in gene regulation. Biochim. Biophys. Acta 1789, 17–25 (2009).
Song, S. H., Hou, C. & Dean, A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell 28, 810–822 (2007).
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012).
Deng, W. et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158, 849–860 (2014). Deng et al. (2012, 2014) demonstrate that ZF-induced enhancer–promoter loop formation can activate transcription of endogenous globin genes.
Oehler, S., Eismann, E. R., Kramer, H. & Muller-Hill, B. The three operators of the lac operon cooperate in repression. EMBO J. 9, 973–979 (1990).
Mandal, N., Su, W., Haber, R., Adhya, S. & Echols, H. DNA looping in cellular repression of transcription of the galactose operon. Genes Dev. 4, 410–418 (1990).
Reitzer, L. J. & Magasanik, B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45, 785–792 (1986).
Priest, D. G. et al. Quantitation of the DNA tethering effect in long-range DNA looping in vivo and in vitro using the Lac and lambda repressors. Proc. Natl Acad. Sci. USA 111, 349–354 (2014).
Cui, L., Murchland, I., Shearwin, K. E. & Dodd, I. B. Enhancer-like long-range transcriptional activation by lambda CI-mediated DNA looping. Proc. Natl Acad. Sci. USA 110, 2922–2927 (2013).
Hao, N., Shearwin, K. E. & Dodd, I. B. Positive and negative control of enhancer-promoter interactions by other DNA loops generates specificity and tunability. Cell Rep. 26, 2419–2433 (2019).
Priest, D. G. et al. Quantitation of interactions between two DNA loops demonstrates loop domain insulation in E. coli cells. Proc. Natl Acad. Sci. USA 111, E4449–E4457 (2014).
Hao, N., Sneppen, K., Shearwin, K. E. & Dodd, I. B. Efficient chromosomal-scale DNA looping in Escherichia coli using multiple DNA-looping elements. Nucleic Acids Res. 45, 5074–5085 (2017).
Breda, L. et al. Forced chromatin looping raises fetal hemoglobin in adult sickle cells to higher levels than pharmacologic inducers. Blood 128, 1139–1143 (2016).
Bartman, C. R., Hsu, S. C., Hsiung, C. C., Raj, A. & Blobel, G. A. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell 62, 237–247 (2016).
Hao, N., Shearwin, K. E. & Dodd, I. B. Programmable DNA looping using engineered bivalent dCas9 complexes. Nat. Commun. 8, 1628 (2017).
Moll, J. R., Ruvinov, S. B., Pastan, I. & Vinson, C. Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10(-15) M. Protein Sci. 10, 649–655 (2001).
Kim, J. H. et al. LADL: light-activated dynamic looping for endogenous gene expression control. Nat. Methods 16, 633–639 (2019). This study develops a programmable, light-activated dynamic looping technique to induce loop formation between two genomic loci targeted by dCas9 proteins, which can induce loop formation to regulate gene expression.
Liang, F. S., Ho, W. Q. & Crabtree, G. R. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal. 4, rs2 (2011).
Kennedy, M. J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010).
Sivakumar, A., de Las Heras, J. I. & Schirmer, E. C. Spatial genome organization: from development to disease. Front. Cell Dev. Biol. 7, 18 (2019).
Meaburn, K. J., Gudla, P. R., Khan, S., Lockett, S. J. & Misteli, T. Disease-specific gene repositioning in breast cancer. J. Cell Biol. 187, 801–812 (2009).
Hogan, M. S., Parfitt, D. E., Zepeda-Mendoza, C. J., Shen, M. M. & Spector, D. L. Transient pairing of homologous Oct4 alleles accompanies the onset of embryonic stem cell differentiation. Cell Stem Cell 16, 275–288 (2015).
Chen, C. K. et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472 (2016).
Andrulis, E. D., Neiman, A. M., Zappulla, D. C. & Sternglanz, R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394, 592–595 (1998).
Taddei, A. et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774–778 (2006).
Reddy, K. L., Zullo, J. M., Bertolino, E. & Singh, H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 (2008).
Kumaran, R. I. & Spector, D. L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180, 51–65 (2008).
Finlan, L. E. et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4, e1000039 (2008). Reddy et al. (2008), Kumaran and Spector (2008) and Finlan et al. (2008) develop lacO–LacI-based systems to reposition the genomic region containing an integrated lacO array to the nuclear periphery.
Zullo, J. M. et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487 (2012).
Ruault, M., Dubarry, M. & Taddei, A. Re-positioning genes to the nuclear envelope in mammalian cells: impact on transcription. Trends Genet. 24, 574–581 (2008).
Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004).
Pollex, T. & Heard, E. Nuclear positioning and pairing of X-chromosome inactivation centers are not primary determinants during initiation of random X-inactivation. Nat. Genet. 51, 285–295 (2019).
Lin, J. L., Ekas, H., Deaner, M. & Alper, H. S. CRISPR-PIN: Modifying gene position in the nucleus via dCas9-mediated tethering. Synth. Syst. Biotechnol. 4, 73–78 (2019).
Wang, H. et al. CRISPR-mediated live imaging of genome editing and transcription. Science 365, 1301–1305 (2019).
Chambeyron, S. & Bickmore, W. A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 (2004).
van Steensel, B. & Furlong, E. E. M. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 20, 327–337 (2019).
Isoda, T. et al. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171, 103–119 (2017).
Tumbar, T. & Belmont, A. S. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 3, 134–139 (2001).
Chuang, C. H. et al. Long-range directional movement of an interphase chromosome site. Curr. Biol. 16, 825–831 (2006).
Therizols, P. et al. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346, 1238–1242 (2014).
Brueckner, L. et al. Local rewiring of genome-nuclear lamina interactions by transcription. EMBO J. 39, e103159 (2020).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Stanek, D. & Fox, A. H. Nuclear bodies: news insights into structure and function. Curr. Opin. Cell Biol. 46, 94–101 (2017).
Wang, Q. et al. Cajal bodies are linked to genome conformation. Nat. Commun. 7, 10966 (2016).
Machyna, M., Neugebauer, K. M. & Stanek, D. Coilin: the first 25 years. RNA Biol. 12, 590–596 (2015).
Zhu, L. & Brangwynne, C. P. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 34, 23–30 (2015).
Berry, J., Weber, S. C., Vaidya, N., Haataja, M. & Brangwynne, C. P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl Acad. Sci. USA 112, E5237–E5245 (2015).
Brangwynne, C. P., Mitchison, T. J. & Hyman, A. A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA 108, 4334–4339 (2011).
Kaiser, T. E., Intine, R. V. & Dundr, M. De novo formation of a subnuclear body. Science 322, 1713–1717 (2008).
Shevtsov, S. P. & Dundr, M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173 (2011).
Chung, I., Leonhardt, H. & Rippe, K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J. Cell Sci. 124, 3603–3618 (2011). Kaiser et al. (2008), Shevtsov and Dundr (2011) and Chung et al. (2011) describe lacO–LacI methods to induce de novo nuclear body formation at the integrated lacO loci by recruiting key structural proteins or RNA components.
Caudron-Herger, M. et al. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 34, 2758–2774 (2015).
Mao, Y. S., Sunwoo, H., Zhang, B. & Spector, D. L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13, 95–101 (2011).
Chujo, T. & Hirose, T. Nuclear bodies built on architectural long noncoding RNAs: unifying principles of their construction and function. Mol. Cell 40, 889–896 (2017).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science https://doi.org/10.1126/science.aar3958 (2018).
Kwon, I. et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013).
Cho, W. K. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018).
Larson, A. G. et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Sanulli, S. et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 (2019).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Burke, K. A., Janke, A. M., Rhine, C. L. & Fawzi, N. L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 (2015).
Sabari, B. R., Dall’Agnese, A. & Young, R. A. Biomolecular condensates in the nucleus. Trends Biochem. Sci. https://doi.org/10.1016/j.tibs.2020.06.007 (2020).
McSwiggen, D. T., Mir, M., Darzacq, X. & Tjian, R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 33, 1619–1634 (2019).
A, P. & Weber, S. C. Evidence for and against liquid-liquid phase separation in the nucleus. Noncoding RNA https://doi.org/10.3390/ncrna5040050 (2019).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171 (2017).
Chong, S. et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science https://doi.org/10.1126/science.aar2555 (2018). This article describes a lacO–LacI-based method to recruit LCDs of FET family proteins (FUS, EWS and TAF15) to form interaction hubs at lacO loci.
Wan, L. et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577, 121–126 (2020).
Erdel, F. et al. Mouse heterochromatin adopts digital compaction states without showing Hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell 78, 236–249 (2020).
Shin, Y. et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 176, 1518 (2019). This article presents the blue light-inducible CasDrop technology to control the formation of phase-separated nuclear condensates at specific genomic loci targeted by dCas9.
Guntas, G. et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl Acad. Sci. USA 112, 112–117 (2015).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016).
Tarjan, D. R., Flavahan, W. A. & Bernstein, B. E. Epigenome editing strategies for the functional annotation of CTCF insulators. Nat. Commun. 10, 4258 (2019).
Acknowledgements
L.S.Q. acknowledges support from the Pew Charitable Trusts, the Alfred P. Sloan Foundation and the Li Ka Shing Foundation. This work was partly supported by US National Institutes of Health Common Fund 4D Nucleome Program (U01 EB021240, 1U01DK127405-01).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Genetics thanks E. Nora and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Nuclear lamina
-
A structural filament meshwork located near the inner nuclear membrane that is composed of lamins and lamin-binding proteins that are associated with membrane proteins and chromatin.
- Enhancer
-
A cis-regulatory DNA element that can bind to transcription factors to promote gene transcription and that may be located close to or far from its associated gene and promoter.
- Promoter
-
A DNA element that can bind to transcription factors and RNA polymerase to initiate gene transcription. Promoters are typically located upstream of the gene that they regulate.
- Chromosome conformation capture (3C)-based techniques
-
A set of techniques to study 3D genome organization by mapping the interaction frequency of genomic loci in three dimensions, including many technical variants such as 3C, 4C, 5C and Hi-C.
- Fluorescence in situ hybridization
-
(FISH). An imaging technique to detect sequence-specific DNA or RNA localization in situ using fluorescently labelled oligonucleotide probes complementary to the imaged DNA or RNA.
- CRISPR live-cell imaging
-
A technique to visualize the dynamics of specific genomic loci or RNA in living cells using CRISPR systems to enrich fluorescent components at the target DNA or RNA.
- Lac operator–Lac repressor
-
(lacO–LacI). lacO, the operator of the bacterial Lac operon, binds tightly to the LacI protein, which represses gene transcription. Binding of isopropyl β-d-1-thiogalactopyranoside to LacI dissociates LacI from lacO.
- Chromosome territory
-
The distinct volume of nuclear space occupied by each chromosome.
- Heterochromatin
-
The condensed region of chromatin that is tightly packaged, which is usually gene-poor. Heterochromatin contains repressive epigenetic markers and is less accessible to transcription factors and transcription machines than is euchromatin.
- Euchromatin
-
The less-condensed region of chromatin that is lightly packaged. Euchromatin is enriched in actively transcribed genes and more accessible to transcription factories than is heterochromatin.
- Nuclear bodies
-
A collection of membraneless structures in the nucleus with diverse cellular functions. Different nuclear bodies are enriched in different structural proteins and RNA components.
- Lamina-associated domains
-
The regions of chromatin that interact with nuclear lamina at the nuclear periphery. They usually contain inactive genes and can be mapped by sequencing approaches such as DamID.
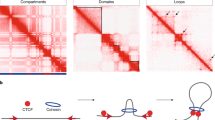
- Topologically associating domains
-
(TADs). Local self-interacting chromatin regions that are often separated by boundary elements. DNA elements exhibit higher frequency of contacts with regions within the same TAD than interactions among TADs.
- Loop
-
A DNA loop is formed when two genome regions in the same chromosome contact, which can bring two distal DNA sites together, mediating interactions between cis-regulatory elements such as promoters, enhancers, insulators and silencers.
- CTCF
-
(CCCTC-binding factor). A conserved 11-zinc-finger protein encoded by the CTCF gene that binds to the consensus DNA sequence CCGCGNGGNGGCAG.
- Cohesin complex
-
A protein complex composed of four core subunits, SMC1, SMC3, RAD21 and SA1 or SA2. It can form a ring-shaped structure enclosing DNA and plays important roles in mitotic and interphase chromosome organization.
- CTCF-binding sites
-
(CBSs). The binding sites of CCCTC-binding factor (CTCF) in the genome that contribute to the organization of topologically associating domains and loops.
- Insulators
-
Cis-regulatory DNA elements that block inappropriate interactions between genomic regions.
- Superloops
-
Networks of super-large loops that exist on the inactive X chromosome around the DXZ4 locus.
- Optogenetic system
-
A light-inducible system that uses light-sensitive proteins to control downstream biological reactions in cells and in vivo.
- Superenhancer
-
A cluster of enhancers that binds to a high level of transcription regulators and mediator complexes. Superenhancers are typically cell-type specific and can control the expression of cell-identity genes.
- Liquid–liquid phase separation
-
(LLPS). A physical process that involves concentrated solutions of polymers (such as proteins and nucleic acids) separating into multiple aqueous phases, which can condense components into liquid droplets.
- Intrinsically disordered regions
-
(IDRs). Proteins or protein segments that do not fold into a defined tertiary structure but can flexibly switch between different conformational states.
- Low-complexity domains
-
(LCDs). Regions in a protein sequence that tend to form flexible and disordered structures, which may contain short amino acid repeats.
Rights and permissions
About this article
Cite this article
Wang, H., Han, M. & Qi, L.S. Engineering 3D genome organization. Nat Rev Genet 22, 343–360 (2021). https://doi.org/10.1038/s41576-020-00325-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-020-00325-5
This article is cited by
-
DiffDomain enables identification of structurally reorganized topologically associating domains
Nature Communications (2024)
-
DNA polymerases in precise and predictable CRISPR/Cas9-mediated chromosomal rearrangements
BMC Biology (2023)
-
Toward cis-regulation in soybean: a 3D genome scope
Molecular Breeding (2023)
-
The 3D architecture of the pepper genome and its relationship to function and evolution
Nature Communications (2022)