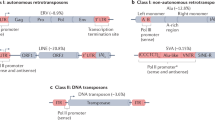
Abstract
Maintenance of genome stability requires control over the expression of transposable elements (TEs), whose activity can have substantial deleterious effects on the host. Chemical modification of DNA is a commonly used strategy to achieve this, and it has long been argued that the emergence of 5-methylcytosine (5mC) in many species was driven by the requirement to silence TEs. Potential roles in TE regulation have also been suggested for other DNA modifications, such as N6-methyladenine and oxidation derivatives of 5mC, although the underlying mechanistic relationships are poorly understood. Here, we discuss current evidence implicating DNA modifications and DNA-modifying enzymes in TE regulation across different species.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
20 March 2019
The originally published article contained an error in Figure 2a: for the left side of the figure part (showing piRNA-directed DNA methylation of mouse transposable elements), DNMT3A/B should have been DNMT3C. The article has now been corrected online.
References
Gregory, T. R. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 76, 65–101 (2001).
Jurka, J., Bao, W. & Kojima, K. K. Families of transposable elements, population structure and the origin of species. Biol. Direct 6, 44 (2011).
Sotero-Caio, C. G., Platt, R. N., Suh, A. & Ray, D. A. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol. Evol. 9, 161–177 (2017).
Feschotte, C. & Betrán, E. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 33, 817–831 (2017).
Joly-Lopez, Z. & Bureau, T. E. Exaptation of transposable element coding sequences. Curr. Opin. Genet. Dev. 49, 34–42 (2018).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017).
Arkhipova, I. R. Neutral theory, transposable elements, and eukaryotic genome evolution. Mol. Biol. Evol. 35, 1332–1337 (2018).
Gilbert, C. & Feschotte, C. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr. Opin. Genet. Dev. 49, 15–24 (2018).
Wicker, T. et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007). This paper presents a comprehensive description of TE classification and nomenclature, based on a combination of TE sequence structure, phylogeny and mechanisms of transposition.
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Kojima, K. K. Human transposable elements in Repbase: genomic footprints from fish to humans. Mob. DNA 9, 2 (2018).
Feschotte, C. & Pritham, E. J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 (2007).
Rodriguez-Terrones, D. & Torres-Padilla, M.-E. Nimble and ready to mingle: transposon outbursts of early development. Trends Genet. 34, 806–820 (2018).
Tsukahara, S. et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature 461, 423–426 (2009).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Richardson, S. R. et al. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res. 27, 1395–1405 (2017).
Brouha, B. et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA 100, 5280–5285 (2003).
Dewannieux, M., Esnault, C. & Heidmann, T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48 (2003).
Hancks, D. C. & Kazazian, H. H. Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9 (2016).
Czech, B. & Hannon, G. J. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 41, 324–337 (2016).
Molaro, A. & Malik, H. S. Hide and seek: how chromatin-based pathways silence retroelements in the mammalian germline. Curr. Opin. Genet. Dev. 37, 51–58 (2016).
Kim, M. Y. & Zilberman, D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 19, 320–326 (2014).
Jacobs, F. M. J. et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245 (2014).
Imbeault, M., Helleboid, P.-Y. & Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017).
Rowe, H. M. & Trono, D. Dynamic control of endogenous retroviruses during development. Virology 411, 273–287 (2011).
Dunican, D. S. et al. Lsh regulates LTR retrotransposon repression independently of Dnmt3b function. Genome Biol. 14, R146 (2013).
Zemach, A. et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205 (2013).
Yoder, J. A., Walsh, C. P. & Bestor, T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997).
Ratel, D., Ravanat, J.-L., Berger, F. & Wion, D. N6-methyladenine: the other methylated base of DNA. Bioessays 28, 309–315 (2006).
Zhang, G. et al. N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906 (2015). This study describes 6mA dynamics during D. melanogaster embryogenesis and reports a correlation between 6mA demethylation and TE suppression.
Wu, T. P. et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 532, 329–333 (2016). This paper is the first to find 6mA in mammalian genomes, identifying both 6mA and its associated demethylase in mouse ESCs, which when removed led to 6mA enrichment at young LINE-1 elements.
O’Brown, Z. K. & Greer, E. L. N6-methyladenine: a conserved and dynamic DNA mark. Adv. Exp. Med. Biol. 945, 213–246 (2016).
Schiffers, S. et al. Quantitative LC-MS provides no evidence for m6dA or m4dC in the genome of mouse embryonic stem cells and tissues. Angew. Chemie 56, 11268–11271 (2017).
Lentini, A. et al. A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat. Methods 15, 499–504 (2018).
Kang, J. et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc. Natl Acad. Sci. USA 112, E4236–E4245 (2015).
la Rica de, L. et al. TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol. 17, 234 (2016). In this paper, the authors show that TET enzymes demethylate LINE-1 elements in ESCs but also recruit the co-repressor SIN3A to ensure LINE-1 silencing.
Zhang, P. et al. L1 retrotransposition is activated by Ten-eleven-translocation protein 1 and repressed by methyl-CpG binding proteins. Nucleus 8, 548–562 (2017).
Deniz, O., la Rica, de, L., Cheng, K. C. L., Spensberger, D. & Branco, M. R. SETDB1 prevents TET2-dependent activation of IAP retroelements in naïve embryonic stem cells. Genome Biol. 19, 6 (2018).
Coluccio, A. et al. Individual retrotransposon integrants are differentially controlled by KZFP/KAP1-dependent histone methylation, DNA methylation and TET-mediated hydroxymethylation in naïve embryonic stem cells. Epigenetics Chromatin 11, 7 (2018).
Du, J., Johnson, L. M., Jacobsen, S. E. & Patel, D. J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 (2015).
Schübeler, D. Function and information content of DNA methylation. Nature 517, 321–326 (2015).
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Rasmussen, K. D. & Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 (2016).
Wu, X. & Zhang, Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017).
Luo, G.-Z. & He, C. DNA N6-methyladenine in metazoans: functional epigenetic mark or bystander? Nat. Struct. Mol. Biol. 24, 503–506 (2017).
Jeltsch, A. Molecular biology. Phylogeny of methylomes. Science 328, 837–838 (2010).
Lechner, M. et al. The correlation of genome size and DNA methylation rate in metazoans. Theory Biosci. 132, 47–60 (2013).
Rošic, S. et al. Evolutionary analysis indicates that DNA alkylation damage is a byproduct of cytosine DNA methyltransferase activity. Nat. Genet. 50, 452–459 (2018).
Lippman, Z., May, B., Yordan, C., Singer, T. & Martienssen, R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLOS Biol. 1, E67 (2003).
Hosaka, A. et al. Evolution of sequence-specific anti-silencing systems in Arabidopsis. Nat. Commun. 8, 2161 (2017).
Zhou, Y., Cambareri, E. B. & Kinsey, J. A. DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol. Genet. Genomics 265, 748–754 (2001).
Chernyavskaya, Y. et al. Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling. Development 144, 2925–2939 (2017).
Walsh, C. P., Chaillet, J. R. & Bestor, T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 (1998). This study is the first to demonstrate the role of DNA methylation in the silencing of TEs (IAPs) in mouse development.
Hutnick, L. K. et al. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum. Mol. Genet. 18, 2875–2888 (2009).
Jackson-Grusby, L. et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27, 31–39 (2001).
Hutnick, L. K., Huang, X., Loo, T.-C., Ma, Z. & Fan, G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J. Biol. Chem. 285, 21082–21091 (2010).
Matsui, T. et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010).
Karimi, M. M. et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687 (2011). This paper demonstrates that DNA methylation and H3K9me3 are targeted to different loci and that SETDB1-mediated H3K9me3 enrichment contributes to the silencing of certain ERVs in mouse ESCs.
Rowe, H. M. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010).
Fasching, L. et al. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Rep. 10, 20–28 (2015).
Castro-Diaz, N. et al. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 28, 1397–1409 (2014).
Molaro, A. et al. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 28, 1544–1549 (2014).
Fadloun, A. et al. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 20, 332–338 (2013). This study reveals the dynamic nature of TE expression during mouse pre-implantation, underlining a transient expression of LINE-1s during this period.
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008).
Hackett, J. A. et al. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development 139, 3623–3632 (2012).
Bourc’his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004). This seminal paper shows in vivo that DNA methylation is required for transposon silencing during spermatogenesis in mice.
Manakov, S. A. et al. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep. 12, 1234–1243 (2015).
Barau, J. et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912 (2016). This study discovers DNMT3C, a fourth DNA methyltransferase enzyme that specifically methylates young TEs in the male germ line.
Jain, D. et al. rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLOS Genet. 13, e1006964 (2017).
Zamudio, N. et al. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256–1270 (2015). In this paper, the authors show that TE silencing during spermatogenesis is required during meiosis owing to an aberrant chromatin structure formed at expressed TE loci, which form meiotic hot spots.
Murchison, E. P. et al. Critical roles for Dicer in the female germline. Genes Dev. 21, 682–693 (2007).
Tam, O. H. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538 (2008).
Kabayama, Y. et al. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res. 45, 5387–5398 (2017).
Malki, S., van der Heijden, G. W., O’Donnell, K. A., Martin, S. L. & Bortvin, A. A. Role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell 29, 521–533 (2014).
Seisenberger, S. et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 (2012). In this paper, the authors describe global DNA methylation dynamics in mouse PGCs featuring DNA methylation-resistant genomic regions, including IAPs, ERV1 and ERVK families.
Lane, N. et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35, 88–93 (2003).
Kobayashi, H. et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 23, 616–627 (2013).
Smith, Z. D. et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 (2012).
Liu, S. et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 28, 2041–2055 (2014). This study identifies SETDB1 as responsible for silencing of DNA demethylation-resistant TEs in PGCs.
Habibi, E. et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13, 360–369 (2013).
von Meyenn, F. et al. Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol. Cell 62, 848–861 (2016). This study shows that replication-dependent passive demethylation is the dominant process during the remodelling of ESC to a naive state. The authors also link H3K9me2 enrichment with UHRF1 recruitment.
Rothbart, S. B. et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160 (2012).
Liu, X. et al. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 4, 1563 (2013).
Maenohara, S. et al. Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLOS Genet. 13, e1007042 (2017).
Walter, M., Teissandier, A., Pérez-Palacios, R. & Bourc’his, D. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. eLife 5, e11418 (2016).
von Meyenn, F. et al. Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Dev. Cell 39, 104–115 (2016).
Sharif, J. et al. Activation of endogenous retroviruses in Dnmt1(−/−) ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell 19, 81–94 (2016).
Berrens, R. V. et al. An endosiRNA-based repression mechanism counteracts transposon activation during global DNA demethylation in embryonic stem cells. Cell Stem Cell 21, 694–703 (2017).
Gaudet, F. et al. Induction of tumors in mice by genomic hypomethylation. Science 300, 489–492 (2003).
Iskow, R. C. et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261 (2010).
Schauer, S. N. et al. L1 retrotransposition is a common feature of mammalian hepatocarcinogenesis. Genome Res. 28, 639–653 (2018).
Nguyen, T. H. M. et al. L1 retrotransposon heterogeneity in ovarian tumor cell evolution. Cell Rep. 23, 3730–3740 (2018).
Rodic, N. et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280–1286 (2014).
Lee, E. et al. Landscape of somatic retrotransposition in human cancers. Science 337, 967–971 (2012). This study provides a detailed overview of somatic TE retrotransposition activity in different types of cancer.
Burns, K. H. Transposable elements in cancer. Nat. Rev. Genet. 17, 415–424 (2017).
Babaian, A. & Mager, D. L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 7, 24 (2016).
Weber, B., Kimhi, S., Howard, G., Eden, A. & Lyko, F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29, 5775–5784 (2010).
Cruickshanks, H. A. & Tufarelli, C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 94, 397–406 (2009).
Brocks, D. et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat. Genet. 49, 1052–1060 (2017).
Cuellar, T. L. et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell Biol. 216, 3535–3549 (2017).
Sheng, W. et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563 (2018).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015). This study is the first to report that tumour-suppressive strategies of DNA-demethylating agents are actually via an interferon response associated with ERV activation.
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Ohtani, H., Liu, M., Zhou, W., Liang, G. & Jones, P. A. Switching roles for DNA and histone methylation depend on evolutionary ages of human endogenous retroviruses. Genome Res. 28, 1147–1157 (2018).
Li, Y., Kumar, S. & Qian, W. Active DNA demethylation: mechanism and role in plant development. Plant Cell Rep. 37, 77–85 (2018).
Wyatt, G. R. & Cohen, S. S. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem. J. 55, 774–782 (1953).
Penn, N. W., Suwalski, R., O’Riley, C., Bojanowski, K. & Yura, R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 126, 781–790 (1972).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009). This study discovers that TET proteins catalyse the conversion of 5mC to 5hmC by an oxidation reaction.
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
He, Y.-F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Inoue, A. & Zhang, Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194–194 (2011).
Hashimoto, H. et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40, 4841–4849 (2012).
Globisch, D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLOS ONE 5, e15367 (2010).
Almeida, R. D. et al. Semi-quantitative immunohistochemical detection of 5-hydroxymethyl-cytosine reveals conservation of its tissue distribution between amphibians and mammals. Epigenetics 7, 137–140 (2012).
Kamstra, J. H., Løken, M., Aleström, P. & Legler, J. Dynamics of DNA hydroxymethylation in zebrafish. Zebrafish 12, 230–237 (2015).
Upton, K. R. et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161, 228–239 (2015).
Szwagierczak, A., Bultmann, S., Schmidt, C. S., Spada, F. & Leonhardt, H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 38, e181 (2010).
Jin, S.-G. et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 71, 7360–7365 (2011).
Pfaffeneder, T. et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chemie 50, 7008–7012 (2011).
Iyer, L. M., Tahiliani, M., Rao, A. & Aravind, L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 8, 1698–1710 (2009).
Iyer, L. M. et al. Lineage-specific expansions of TET/JBP genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes. Proc. Natl Acad. Sci. USA 111, 1676–1683 (2014).
Chavez, L. et al. Simultaneous sequencing of oxidized methylcytosines produced by TET/JBP dioxygenases in Coprinopsis cinerea. Proc. Natl Acad. Sci. USA 111, E5149–E5158 (2014).
Wang, X.-L. et al. Genome-wide mapping of 5-hydroxymethylcytosine in three rice cultivars reveals its preferential localization in transcriptionally silent transposable element genes. J. Exp. Bot. 66, 6651–6663 (2015).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011).
Booth, M. J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Inoue, A., Shen, L., Dai, Q., He, C. & Zhang, Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 21, 1670–1676 (2011).
Gu, T.-P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011).
Amouroux, R. et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233 (2016).
Shen, L. et al. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15, 459–470 (2014).
Kim, S.-H. et al. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem. Biophys. Res. Commun. 324, 58–63 (2004).
Inoue, A., Matoba, S. & Zhang, Y. Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation. Cell Res. 22, 1640–1649 (2012).
Vella, P. et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 (2013).
Chen, Q., Chen, Y., Bian, C., Fujiki, R. & Yu, X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 (2013).
Deplus, R. et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 (2013).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Neri, F. et al. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 14, R91 (2013).
Guallar, D. et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 30, 733 (2018).
Leung, D. et al. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc. Natl Acad. Sci. USA 111, 6690–6695 (2014).
Bachman, M. et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6, 1049–1055 (2014).
Bachman, M. et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 11, 555–557 (2015).
Hashimoto, H. et al. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 28, 2304–2313 (2014).
Kellinger, M. W. et al. 5-Formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 19, 831–833 (2012).
Iurlaro, M. et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14, R119 (2013).
Spruijt, C. G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013).
Xiong, J. et al. Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol. Cell 64, 913–925 (2016).
Fu, Y. et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892 (2015).
Greer, E. L. et al. DNA methylation on N6-adenine in C. elegans. Cell 161, 868–878 (2015).
Koziol, M. J. et al. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 23, 24–30 (2016).
Xiao, C.-L. et al. N6-methyladenine DNA modification in the human genome. Mol. Cell 71, 306–318 (2018).
Sánchez-Romero, M. A., Cota, I. & Casadesús, J. DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 25, 9–16 (2015).
Roberts, D., Hoopes, B. C., McClure, W. R. & Kleckner, N. IS10 transposition is regulated by DNA adenine methylation. Cell 43, 117–130 (1985). Dam -mutant E. coli are used to show that 6mA loss results in increased transcription of the IS10 transposon and that this leads to transposition.
Wang, Y., Chen, X., Sheng, Y., Liu, Y. & Gao, S. N6-adenine DNA methylation is associated with the linker DNA of H2A. Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 45, 11594–11606 (2017).
Chen, H. et al. Phytophthora methylomes are modulated by 6 mA methyltransferases and associated with adaptive genome regions. Genome Biol. 19, 181 (2018).
Liang, Z. et al. DNA N6-adenine methylation in Arabidopsis thaliana. Dev. Cell 45, 406–416 (2018).
Liu, J. et al. Abundant DNA 6 mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 7, 13052 (2016).
Yao, B. et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 8, 1122 (2017).
Zhu, S. et al. Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 28, 1067–1078 (2018).
Xie, Q. et al. N6-methyladenine DNA modification in glioblastoma. Cell 175, 1228–1243 (2018).
Mondo, S. J. et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 49, 964–968 (2017).
Brocken, D. J. W., Tark-Dame, M. & Dame, R. T. dCas9: a versatile tool for epigenome editing. Curr. Issues Mol. Biol. 26, 15–32 (2018).
Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Zhao, B. S., Roundtree, I. A. & He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Wan, Y. et al. Transcriptome-wide high-throughput deep m(6)A-seq reveals unique differential m(6)A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 16, 272 (2015).
Zhang, Z. & Xing, Y. CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome. Nucleic Acids Res. 45, 9260–9271 (2017).
Huang, L., Ashraf, S., Wang, J. & Lilley, D. M. Control of box C/D snoRNP assembly by N6-methylation of adenine. EMBO Rep. 18, 1631–1645 (2017).
Zhou, C. et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20, 2262–2276 (2017).
Horwich, M. D. et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272 (2007).
Saito, K. et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 21, 1603–1608 (2007).
Kamminga, L. M. et al. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 29, 3688–3700 (2010).
Jackman, J. E. & Alfonzo, J. D. Transfer RNA modifications: nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 4, 35–48 (2013).
Chou, H.-J., Donnard, E., Gustafsson, H. T., Garber, M. & Rando, O. J. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol. Cell 68, 978–992 (2017).
Phalke, S. et al. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 41, 696–702 (2009).
Kuhlmann, M. et al. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res. 33, 6405–6417 (2005).
Raddatz, G. et al. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl Acad. Sci. USA 110, 8627–8631 (2013).
Genenncher, B. et al. Mutations in cytosine-5 tRNA methyltransferases impact mobile element expression and genome stability at specific DNA repeats. Cell Rep. 22, 1861–1874 (2018).
Ibarra, C. A. et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364 (2012). This paper provides genome-wide evidence that flowering plants use companion cells to protect their gametes from harmful transposition.
Rowe, H. M. et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140, 519–529 (2013).
Acknowledgements
The authors thank M. Lorincz, R. Meehan and the reviewers for detailed comments on the manuscript, which led to extensive improvements to this Review. They also thank C. Feschotte for input on figure 1. They apologize to colleagues whose work was not cited owing to space limitations. M.R.B. is a Sir Henry Dale Fellow (101225/Z/13/Z), jointly funded by the Wellcome Trust and the Royal Society. Ö.D. has received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under REA grant agreement number 608765.
Reviewer information
Nature Reviews Genetics thanks J. Dejardin, M. Gauchier, J. Pontis, D. Trono and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
All authors contributed to all aspects of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Genetic drift
-
The changes in the frequency of a given allele in a population due to random sampling. Genetic drift can lead to the fixation of a particular allelic variant in a population without any selective pressure.
- Horizontal propagation
-
Better known as horizontal gene transfer, horizontal propagation entails the transfer of genetic material between organisms. It contrasts with vertical transfer, which occurs from parents to offspring via the germline.
- PIWI-interacting RNAs
-
(piRNAs). A class of small, single-stranded RNAs of 26–30 nucleotides that interacts with the PIWI family of proteins.
- Post-transcriptional gene silencing
-
(PTGS). The process of silencing a gene after it has been transcribed, for example, by cleavage of its nascent RNA.
- Transcriptional gene silencing
-
The silencing of a gene at the transcriptional level, that is, by preventing the transcriptional process, often by epigenetic modification of the locus to a less open conformation, disfavouring binding of RNA polymerase II.
- RNA-dependent DNA methylation
-
(RdDM). One of the key strategies for de novo and maintenance DNA methylation in Arabidopsis thaliana, whereby RNA molecules from expressed loci direct DNA methylation in a sequence-dependent manner.
- Pre-implantation development
-
The first phase of embryonic development that begins after fertilization and ends upon implantation of the blastocyst into the uterus.
- Endogenous small interfering RNAs
-
(endosiRNAs). Small RNAs (20–23 nucleotides) generated from double-stranded RNAs, including sense–antisense transcript hybrids.
- Primordial germ cells
-
(PGCs). The precursor cells of mammalian gametes that are specified at approximately embryonic day 6.25 in mice and that differentiate into oocytes or sperm.
- Naive pluripotency
-
A stem cell state that resembles that of the inner cell mass of the blastocyst.
- Chimeric transcripts
-
In the context of this Review, chimeric transcripts are RNA molecules that involve a fusion between a transposable element acting as a transcriptional promoter and a host gene.
- Clonal selection
-
In the context of cancer evolution, clonal selection entails the selective expansion of a particular cell due to genetic and/or epigenetic changes that confer a growth advantage.
- Zygotes
-
One-cell embryos resulting from the fusion of sperm with an oocyte, that is, fertilization.
- Orthologue
-
A gene from different species that has evolved from a common ancestor.
Rights and permissions
About this article
Cite this article
Deniz, Ö., Frost, J.M. & Branco, M.R. Regulation of transposable elements by DNA modifications. Nat Rev Genet 20, 417–431 (2019). https://doi.org/10.1038/s41576-019-0106-6
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-019-0106-6
This article is cited by
-
DNA methylation haplotype block signatures responding to Staphylococcus aureus subclinical mastitis and association with production and health traits
BMC Biology (2024)
-
BSXplorer: analytical framework for exploratory analysis of BS-seq data
BMC Bioinformatics (2024)
-
Durable and efficient gene silencing in vivo by hit-and-run epigenome editing
Nature (2024)
-
Auto-suppression of Tet dioxygenases protects the mouse oocyte genome from oxidative demethylation
Nature Structural & Molecular Biology (2024)
-
Reading banned regions of genomes
Nature Plants (2024)